Liquid-liquid phase separation of H3K27me3 reader BP1 regulates transcriptional repression
- PMID: 38468348
- PMCID: PMC10926671
- DOI: 10.1186/s13059-024-03209-7
Liquid-liquid phase separation of H3K27me3 reader BP1 regulates transcriptional repression
Abstract
Background: Bromo-adjacent homology-plant homeodomain domain containing protein 1 (BP1) is a reader of histone post-translational modifications in fungi. BP1 recognizes trimethylation of lysine 27 in histone H3 (H3K27me3), an epigenetic hallmark of gene silencing. However, whether and how BP1 participates in transcriptional repression remains poorly understood.
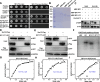
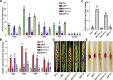
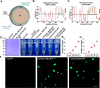
Results: We report that BP1 forms phase-separated liquid condensates to modulate its biological function in Fusarium graminearum. Deletion assays reveal that intrinsically disordered region 2 (IDR2) of BP1 mediates its liquid-liquid phase separation. The phase separation of BP1 is indispensable for its interaction with suppressor of Zeste 12, a component of polycomb repressive complex 2. Furthermore, IDR2 deletion abolishes BP1-H3K27me3 binding and alleviates the transcriptional repression of secondary metabolism-related genes, especially deoxynivalenol mycotoxin biosynthesis genes.
Conclusions: BP1 maintains transcriptional repression by forming liquid-liquid phase-separated condensates, expanding our understanding of the relationship between post-translational modifications and liquid-liquid phase separation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Fusarium BP1 is a reader of H3K27 methylation.Nucleic Acids Res. 2021 Oct 11;49(18):10448-10464. doi: 10.1093/nar/gkab844. Nucleic Acids Res. 2021. PMID: 34570240 Free PMC article.
-
PHF1 compartmentalizes PRC2 via phase separation.Biochem J. 2023 Nov 29;480(22):1833-1844. doi: 10.1042/BCJ20230040. Biochem J. 2023. PMID: 37888776
-
A repressive H3K36me2 reader mediates Polycomb silencing.Nat Commun. 2024 Aug 24;15(1):7287. doi: 10.1038/s41467-024-51789-6. Nat Commun. 2024. PMID: 39179589 Free PMC article.
-
Not just a writer: PRC2 as a chromatin reader.Biochem Soc Trans. 2021 Jun 30;49(3):1159-1170. doi: 10.1042/BST20200728. Biochem Soc Trans. 2021. PMID: 34060617 Free PMC article. Review.
-
A Structural Perspective on Gene Repression by Polycomb Repressive Complex 2.Subcell Biochem. 2021;96:519-562. doi: 10.1007/978-3-030-58971-4_17. Subcell Biochem. 2021. PMID: 33252743 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

