Comparative transcriptome and metabolome profiles of the leaf and fruits of a Xianjinfeng litchi budding mutant and its mother plant
- PMID: 38463170
- PMCID: PMC10920226
- DOI: 10.3389/fgene.2024.1360138
Comparative transcriptome and metabolome profiles of the leaf and fruits of a Xianjinfeng litchi budding mutant and its mother plant
Abstract
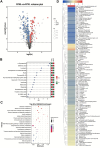
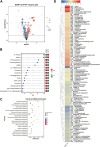
Background: Litchi (Litchi chinensis) is an important sub-tropical fruit in the horticulture market in China. Breeding for improved fruit characteristics is needed for satisfying consumer demands. Budding is a sustainable method for its propagation. During our ongoing breeding program, we observed a litchi mutant with flat leaves and sharp fruit peel cracking in comparison to the curled leaves and blunt fruit peel cracking fruits of the mother plant. Methods: To understand the possible molecular pathways involved, we performed a combined metabolome and transcriptome analysis. Results: We identified 1,060 metabolites in litchi leaves and fruits, of which 106 and 101 were differentially accumulated between the leaves and fruits, respectively. The mutant leaves were richer in carbohydrates, nucleotides, and phenolic acids, while the mother plant was rich in most of the amino acids and derivatives, flavonoids, lipids and organic acids and derivatives, and vitamins. Contrastingly, mutant fruits had higher levels of amino acids and derivatives, carbohydrates and derivatives, and organic acids and derivatives. However, the mother plant's fruits contained higher levels of flavonoids, scopoletin, amines, some amino acids and derivatives, benzamidine, carbohydrates and derivatives, and some organic acids and derivatives. The number of differentially expressed genes was consistent with the metabolome profiles. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway-enriched gene expressions showed consistent profiles as of metabolome analysis. Conclusion: These results provide the groundwork for breeding litchi for fruit and leaf traits that are useful for its taste and yield.
Keywords: amino acids and derivatives; carbohydrates and derivatives; flavonoid metabolome in fruits; leaf folding; litchi fruit.
Copyright © 2024 Xu, Qin, Li, Hou, Fang, Zhang, You, Li and Qiu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
De novo assembly and characterization of pericarp transcriptome and identification of candidate genes mediating fruit cracking in Litchi chinensis Sonn.Int J Mol Sci. 2014 Sep 30;15(10):17667-85. doi: 10.3390/ijms151017667. Int J Mol Sci. 2014. PMID: 25272225 Free PMC article.
-
Combined Metabolome and Transcriptome Analyses Unveil the Molecular Mechanisms of Fruit Acidity Variation in Litchi (Litchi chinensis Sonn.).Int J Mol Sci. 2023 Jan 18;24(3):1871. doi: 10.3390/ijms24031871. Int J Mol Sci. 2023. PMID: 36768192 Free PMC article.
-
Metabolome and transcriptome analysis of terpene synthase genes and their putative role in floral aroma production in Litchi chinensis.Physiol Plant. 2022 Nov;174(6):e13796. doi: 10.1111/ppl.13796. Physiol Plant. 2022. PMID: 36251666
-
A review on the medicinal potential, toxicology, and phytochemistry of litchi fruit peel and seed.J Food Biochem. 2021 Dec;45(12):e13997. doi: 10.1111/jfbc.13997. Epub 2021 Nov 8. J Food Biochem. 2021. PMID: 34750843 Review.
-
[Fruit cracking: a review].Sheng Wu Gong Cheng Xue Bao. 2021 Aug 25;37(8):2737-2752. doi: 10.13345/j.cjb.200553. Sheng Wu Gong Cheng Xue Bao. 2021. PMID: 34472292 Review. Chinese.
References
-
- Bylesjö M., Rantalainen M., Cloarec O., Nicholson J. K., Holmes E., Trygg J. (2006). OPLS discriminant analysis: combining the strengths of PLS‐DA and SIMCA classification. J. Chemom. A J. Chemom. Soc. 20, 341–351. 10.1002/cem.1006 - DOI
-
- Chen H., Wan Y., Teng K., Liu B., Zhao N., Xu K., et al. (2023). The role of OsOFP8 gene in regulating rice leaf angle. J. Plant Biochem. Biotechnol. 32, 304–318. 10.1007/s13562-022-00806-0 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

