Functional analyses of bacterial genomes found in Symbiodiniaceae genome assemblies
- PMID: 38444256
- PMCID: PMC10915500
- DOI: 10.1111/1758-2229.13238
Functional analyses of bacterial genomes found in Symbiodiniaceae genome assemblies
Abstract
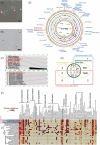
Bacterial-algal interactions strongly influence marine ecosystems. Bacterial communities in cultured dinoflagellates of the family Symbiodiniaceae have been characterized by metagenomics. However, little is known about whole-genome analysis of marine bacteria associated with these dinoflagellates. We performed in silico analysis of four bacterial genomes from cultures of four dinoflagellates of the genera Symbiodinium, Breviolum, Cladocopium and Durusdinium. Comparative analysis showed that the former three contain the alphaproteobacterial family Parvibaculaceae and that the Durusdinium culture includes the family Sphingomonadaceae. There were no large genomic reductions in the alphaproteobacteria with genome sizes of 2.9-3.9 Mb, implying they are not obligate intracellular bacteria. Genomic annotations of three Parvibaculaceae detected the gene for diacetylchitobiose deacetylase (Dac), which may be involved in the degradation of dinoflagellate cell surfaces. They also had metabolic genes for dissimilatory nitrate reduction to ammonium (DNRA) in the nitrogen (N) cycle and cobalamin (vitamin B12 ) biosynthetic genes in the salvage pathway. Those three characters were not found in the Sphingomonadaceae genome. Predicted biosynthetic gene clusters for secondary metabolites indicated that the Parvibaculaceae likely produce the same secondary metabolites. Our study suggests that the Parvibaculaceae is a major resident of Symbiodiniaceae cultures with antibiotics.
© 2024 The Authors. Environmental Microbiology Reports published by Applied Microbiology International and John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures

Similar articles
-
A New Dinoflagellate Genome Illuminates a Conserved Gene Cluster Involved in Sunscreen Biosynthesis.Genome Biol Evol. 2021 Feb 3;13(2):evaa235. doi: 10.1093/gbe/evaa235. Genome Biol Evol. 2021. PMID: 33146374 Free PMC article.
-
Gene clusters for biosynthesis of mycosporine-like amino acids in dinoflagellate nuclear genomes: Possible recent horizontal gene transfer between species of Symbiodiniaceae (Dinophyceae).J Phycol. 2022 Feb;58(1):1-11. doi: 10.1111/jpy.13219. Epub 2021 Nov 26. J Phycol. 2022. PMID: 34699617 Free PMC article. Review.
-
Diversity and distribution of Symbiodiniaceae detected on coral reefs of Lombok, Indonesia using environmental DNA metabarcoding.PeerJ. 2022 Oct 24;10:e14006. doi: 10.7717/peerj.14006. eCollection 2022. PeerJ. 2022. PMID: 36312748 Free PMC article.
-
Ammonia transporter 2 as a molecular marker to elucidate the potentials of ammonia transport in phylotypes of Symbiodinium, Cladocopium and Durusdinium in the fluted giant clam, Tridacna squamosa.Comp Biochem Physiol A Mol Integr Physiol. 2022 Jul;269:111225. doi: 10.1016/j.cbpa.2022.111225. Epub 2022 Apr 20. Comp Biochem Physiol A Mol Integr Physiol. 2022. PMID: 35460895
-
Genome Evolution of Coral Reef Symbionts as Intracellular Residents.Trends Ecol Evol. 2019 Sep;34(9):799-806. doi: 10.1016/j.tree.2019.04.010. Epub 2019 May 10. Trends Ecol Evol. 2019. PMID: 31084944 Review.
References
-
- Amin, S.A. , Hmelo, L.R. , van Tol, H.M. , Durham, B.P. , Carlson, L.T. , Heal, K.R. et al. (2015) Interaction and signaling between a cosmopolitan phytoplankton and associated bacteria. Nature, 522, 98–101. - PubMed
-
- Bernasconi, R. , Stat, M. , Koenders, A. & Huggett, M.J. (2018) Global networks of Symbiodinium‐bacteria within the coral Holobiont. Microbial Ecology, 77, 794–807. - PubMed
-
- Burkholder, P.R. (1973) The ecology of marine antibiotics and coral reefs. In: Jones, O.A. & Endean, R. (Eds.) Biology and geology of coral reefs, vol II. Biology 1. New York: Academic Press, pp. 117–182.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

