Evolution of a Restriction Factor by Domestication of a Yeast Retrotransposon
- PMID: 38442736
- PMCID: PMC10951436
- DOI: 10.1093/molbev/msae050
Evolution of a Restriction Factor by Domestication of a Yeast Retrotransposon
Abstract
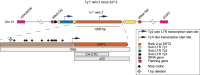
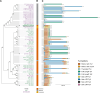
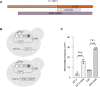
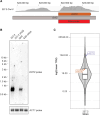
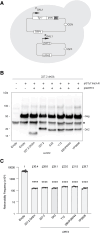
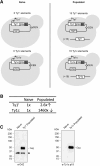
Transposable elements drive genome evolution in all branches of life. Transposable element insertions are often deleterious to their hosts and necessitate evolution of control mechanisms to limit their spread. The long terminal repeat retrotransposon Ty1 prime (Ty1'), a subfamily of the Ty1 family, is present in many Saccharomyces cerevisiae strains, but little is known about what controls its copy number. Here, we provide evidence that a novel gene from an exapted Ty1' sequence, domesticated restriction of Ty1' relic 2 (DRT2), encodes a restriction factor that inhibits Ty1' movement. DRT2 arose through domestication of a Ty1' GAG gene and contains the C-terminal domain of capsid, which in the related Ty1 canonical subfamily functions as a self-encoded restriction factor. Bioinformatic analysis reveals the widespread nature of DRT2, its evolutionary history, and pronounced structural variation at the Ty1' relic 2 locus. Ty1' retromobility analyses demonstrate DRT2 restriction factor functionality, and northern blot and RNA-seq analysis indicate that DRT2 is transcribed in multiple strains. Velocity cosedimentation profiles indicate an association between Drt2 and Ty1' virus-like particles or assembly complexes. Chimeric Ty1' elements containing DRT2 retain retromobility, suggesting an ancestral role of productive Gag C-terminal domain of capsid functionality is present in the sequence. Unlike Ty1 canonical, Ty1' retromobility increases with copy number, suggesting that C-terminal domain of capsid-based restriction is not limited to the Ty1 canonical subfamily self-encoded restriction factor and drove the endogenization of DRT2. The discovery of an exapted Ty1' restriction factor provides insight into the evolution of the Ty1 family, evolutionary hot-spots, and host-transposable element interactions.
Keywords: Saccharomyces; domesticated gene; host defense; retrotransposon.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Evolution of Ty1 copy number control in yeast by horizontal transfer and recombination.PLoS Genet. 2020 Feb 21;16(2):e1008632. doi: 10.1371/journal.pgen.1008632. eCollection 2020 Feb. PLoS Genet. 2020. PMID: 32084126 Free PMC article.
-
A self-encoded capsid derivative restricts Ty1 retrotransposition in Saccharomyces.Curr Genet. 2016 May;62(2):321-9. doi: 10.1007/s00294-015-0550-6. Epub 2015 Dec 9. Curr Genet. 2016. PMID: 26650614 Free PMC article. Review.
-
Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence.Genome Res. 1998 May;8(5):464-78. doi: 10.1101/gr.8.5.464. Genome Res. 1998. PMID: 9582191
-
A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control.J Virol. 2015 Apr;89(7):3922-38. doi: 10.1128/JVI.03060-14. Epub 2015 Jan 21. J Virol. 2015. PMID: 25609815 Free PMC article.
-
The Ty1 LTR-Retrotransposon of Budding Yeast, Saccharomyces cerevisiae.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0053-2014. doi: 10.1128/microbiolspec.MDNA3-0053-2014. Microbiol Spectr. 2015. PMID: 26104690 Review.
References
-
- Basile A, De Pascale F, Bianca F, Rossi A, Frizzarin M, De Bernardini N, Bosaro M, Baldisseri A, Antoniali P, Lopreiato R, et al. Large-scale sequencing and comparative analysis of oenological Saccharomyces cerevisiae strains supported by nanopore refinement of key genomes. Food Microbiol. 2021:97:103753. 10.1016/j.fm.2021.103753. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

