Genomic surveillance of SARS-CoV-2 using long-range PCR primers
- PMID: 38440140
- PMCID: PMC10910555
- DOI: 10.3389/fmicb.2024.1272972
Genomic surveillance of SARS-CoV-2 using long-range PCR primers
Abstract
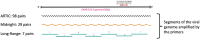
Introduction: Whole Genome Sequencing (WGS) of the SARS-CoV-2 virus is crucial in the surveillance of the COVID-19 pandemic. Several primer schemes have been developed to sequence nearly all of the ~30,000 nucleotide SARS-CoV-2 genome, using a multiplex PCR approach to amplify cDNA copies of the viral genomic RNA. Midnight primers and ARTIC V4.1 primers are the most popular primer schemes that can amplify segments of SARS-CoV-2 (400 bp and 1200 bp, respectively) tiled across the viral RNA genome. Mutations within primer binding sites and primer-primer interactions can result in amplicon dropouts and coverage bias, yielding low-quality genomes with 'Ns' inserted in the missing amplicon regions, causing inaccurate lineage assignments, and making it challenging to monitor lineage-specific mutations in Variants of Concern (VoCs).
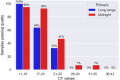
Methods: In this study we used a set of seven long-range PCR primer pairs to sequence clinical isolates of SARS-CoV-2 on Oxford Nanopore sequencer. These long-range primers generate seven amplicons approximately 4500 bp that covered whole genome of SARS-CoV-2. One of these regions includes the full-length S-gene by using a set of flanking primers. We also evaluated the performance of these long-range primers with Midnight primers by sequencing 94 clinical isolates in a Nanopore flow cell.
Results and discussion: Using a small set of long-range primers to sequence SARS-CoV-2 genomes reduces the possibility of amplicon dropout and coverage bias. The key finding of this study is that long range primers can be used in single-molecule sequencing of RNA viruses in surveillance of emerging variants. We also show that by designing primers flanking the S-gene, we can obtain reliable identification of SARS-CoV-2 variants.
Keywords: SARS-CoV-2; genomic epidemiology; long-range primers; nanopore; sequencing; surveillance.
Copyright © 2024 Kandel, Hartzell, Ingold, Turner, Kennedy and Ussery.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


Update of
-
Genomic Surveillance of SARS-CoV-2 Using Long-Range PCR Primers.bioRxiv [Preprint]. 2023 Jul 11:2023.07.10.548464. doi: 10.1101/2023.07.10.548464. bioRxiv. 2023. Update in: Front Microbiol. 2024 Feb 14;15:1272972. doi: 10.3389/fmicb.2024.1272972. PMID: 37502853 Free PMC article. Updated. Preprint.
Similar articles
-
Genomic Surveillance of SARS-CoV-2 Using Long-Range PCR Primers.bioRxiv [Preprint]. 2023 Jul 11:2023.07.10.548464. doi: 10.1101/2023.07.10.548464. bioRxiv. 2023. Update in: Front Microbiol. 2024 Feb 14;15:1272972. doi: 10.3389/fmicb.2024.1272972. PMID: 37502853 Free PMC article. Updated. Preprint.
-
Analysis of the ARTIC V4 and V4.1 SARS-CoV-2 primers and their impact on the detection of Omicron BA.1 and BA.2 lineage-defining mutations.Microb Genom. 2023 Apr;9(4):mgen000991. doi: 10.1099/mgen.0.000991. Microb Genom. 2023. PMID: 37083576 Free PMC article.
-
Universal whole-genome Oxford nanopore sequencing of SARS-CoV-2 using tiled amplicons.Sci Rep. 2023 Jun 26;13(1):10334. doi: 10.1038/s41598-023-37588-x. Sci Rep. 2023. PMID: 37365249 Free PMC article.
-
A short plus long-amplicon based sequencing approach improves genomic coverage and variant detection in the SARS-CoV-2 genome.PLoS One. 2022 Jan 13;17(1):e0261014. doi: 10.1371/journal.pone.0261014. eCollection 2022. PLoS One. 2022. PMID: 35025877 Free PMC article.
-
SARS-CoV-2 Variants Identification: Overview of Molecular Existing Methods.Pathogens. 2022 Sep 17;11(9):1058. doi: 10.3390/pathogens11091058. Pathogens. 2022. PMID: 36145490 Free PMC article. Review.
Cited by
-
Hybrid-Capture Target Enrichment in Human Pathogens: Identification, Evolution, Biosurveillance, and Genomic Epidemiology.Pathogens. 2024 Mar 23;13(4):275. doi: 10.3390/pathogens13040275. Pathogens. 2024. PMID: 38668230 Free PMC article. Review.
-
ViralPrimer: a web server to monitor viral nucleic acid amplification tests' primer efficiency during pandemics, with emphasis on SARS-CoV-2 and Mpox.Bioinformatics. 2024 Nov 1;40(11):btae657. doi: 10.1093/bioinformatics/btae657. Bioinformatics. 2024. PMID: 39499138 Free PMC article.
References
-
- Aksamentov I., Roemer C., Hodcroft E. B., Neher R. A. (2021). Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 6:3773. doi: 10.21105/joss.03773 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

