Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution
- PMID: 38431681
- PMCID: PMC10908815
- DOI: 10.1038/s41467-024-46239-2
Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution
Abstract
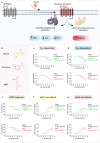
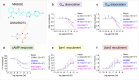
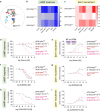
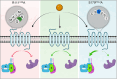
The Hydroxycarboxylic acid receptor 2 (HCA2), also known as the niacin receptor or GPR109A, is a prototypical GPCR that plays a central role in the inhibition of lipolytic and atherogenic activities. Its activation also results in vasodilation that is linked to the side-effect of flushing associated with dyslipidemia drugs such as niacin. GPR109A continues to be a target for developing potential therapeutics in dyslipidemia with minimized flushing response. Here, we present cryo-EM structures of the GPR109A in complex with dyslipidemia drugs, niacin or acipimox, non-flushing agonists, MK6892 or GSK256073, and recently approved psoriasis drug, monomethyl fumarate (MMF). These structures elucidate the binding mechanism of agonists, molecular basis of receptor activation, and insights into biased signaling elicited by some of the agonists. The structural framework also allows us to engineer receptor mutants that exhibit G-protein signaling bias, and therefore, our study may help in structure-guided drug discovery efforts targeting this receptor.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Discovery and characterization of GSK256073, a non-flushing hydroxy-carboxylic acid receptor 2 (HCA2) agonist.Eur J Pharmacol. 2015 Jun 5;756:1-7. doi: 10.1016/j.ejphar.2015.01.051. Epub 2015 Mar 12. Eur J Pharmacol. 2015. PMID: 25773497 Clinical Trial.
-
Differential tissue and ligand-dependent signaling of GPR109A receptor: implications for anti-atherosclerotic therapeutic potential.Cell Signal. 2013 Oct;25(10):2003-16. doi: 10.1016/j.cellsig.2013.06.008. Epub 2013 Jun 14. Cell Signal. 2013. PMID: 23770183
-
Niacin lipid efficacy is independent of both the niacin receptor GPR109A and free fatty acid suppression.Sci Transl Med. 2012 Aug 22;4(148):148ra115. doi: 10.1126/scitranslmed.3003877. Sci Transl Med. 2012. PMID: 22914621 Clinical Trial.
-
Novel Niacin Receptor Agonists: A Promising Strategy for the Treatment of Dyslipidemia.Mini Rev Med Chem. 2021;21(17):2481-2496. doi: 10.2174/1389557521666210125144921. Mini Rev Med Chem. 2021. PMID: 33550969 Review.
-
Future of GPR109A agonists in the treatment of dyslipidaemia.Diabetes Obes Metab. 2011 Aug;13(8):685-91. doi: 10.1111/j.1463-1326.2011.01400.x. Diabetes Obes Metab. 2011. PMID: 21418500 Review.
Cited by
-
Orphan GPCRs in Neurodegenerative Disorders: Integrating Structural Biology and Drug Discovery Approaches.Curr Issues Mol Biol. 2024 Oct 19;46(10):11646-11664. doi: 10.3390/cimb46100691. Curr Issues Mol Biol. 2024. PMID: 39451571 Free PMC article. Review.
-
Multiple recent HCAR2 structures demonstrate a highly dynamic ligand binding and G protein activation mode.Nat Commun. 2024 Jun 25;15(1):5364. doi: 10.1038/s41467-024-49536-y. Nat Commun. 2024. PMID: 38918366 Free PMC article. Review.
-
Molecular mechanism of distinct chemokine engagement and functional divergence of the human Duffy antigen receptor.Cell. 2024 Aug 22;187(17):4751-4769.e25. doi: 10.1016/j.cell.2024.07.005. Epub 2024 Jul 31. Cell. 2024. PMID: 39089252 Free PMC article.
-
Hypertriglyceridemia Therapy: Past, Present and Future Perspectives.Int J Mol Sci. 2024 Sep 8;25(17):9727. doi: 10.3390/ijms25179727. Int J Mol Sci. 2024. PMID: 39273674 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

