Epigenetic Restriction Factors (eRFs) in Virus Infection
- PMID: 38399958
- PMCID: PMC10892949
- DOI: 10.3390/v16020183
Epigenetic Restriction Factors (eRFs) in Virus Infection
Abstract
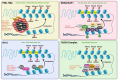
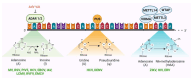
The ongoing arms race between viruses and their hosts is constantly evolving. One of the ways in which cells defend themselves against invading viruses is by using restriction factors (RFs), which are cell-intrinsic antiviral mechanisms that block viral replication and transcription. Recent research has identified a specific group of RFs that belong to the cellular epigenetic machinery and are able to restrict the gene expression of certain viruses. These RFs can be referred to as epigenetic restriction factors or eRFs. In this review, eRFs have been classified into two categories. The first category includes eRFs that target viral chromatin. So far, the identified eRFs in this category include the PML-NBs, the KRAB/KAP1 complex, IFI16, and the HUSH complex. The second category includes eRFs that target viral RNA or, more specifically, the viral epitranscriptome. These epitranscriptomic eRFs have been further classified into two types: those that edit RNA bases-adenosine deaminase acting on RNA (ADAR) and pseudouridine synthases (PUS), and those that covalently modify viral RNA-the N6-methyladenosine (m6A) writers, readers, and erasers. We delve into the molecular machinery of eRFs, their role in limiting various viruses, and the mechanisms by which viruses have evolved to counteract them. We also examine the crosstalk between different eRFs, including the common effectors that connect them. Finally, we explore the potential for new discoveries in the realm of epigenetic networks that restrict viral gene expression, as well as the future research directions in this area.
Keywords: ADAR; HUSH complex; IFI16; KRAB/KAP1; N6 methyl adenosine (m6A); PML-NB; epigenetic viral restriction factor (eRF); epitranscriptomics; pseudouridine synthases (PUS); viral chromatin.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Mechanisms of Host IFI16, PML, and Daxx Protein Restriction of Herpes Simplex Virus 1 Replication.J Virol. 2018 Apr 27;92(10):e00057-18. doi: 10.1128/JVI.00057-18. Print 2018 May 15. J Virol. 2018. PMID: 29491153 Free PMC article.
-
Association of N6-methyladenosine with viruses and related diseases.Virol J. 2019 Nov 11;16(1):133. doi: 10.1186/s12985-019-1236-3. Virol J. 2019. PMID: 31711514 Free PMC article. Review.
-
RNA epitranscriptomics: Regulation of infection of RNA and DNA viruses by N6 -methyladenosine (m6 A).Rev Med Virol. 2018 Jul;28(4):e1983. doi: 10.1002/rmv.1983. Epub 2018 Apr 26. Rev Med Virol. 2018. PMID: 29698584 Free PMC article. Review.
-
Association of N6-methyladenosine with viruses and virally induced diseases.Front Biosci (Landmark Ed). 2020 Mar 1;25(6):1184-1201. doi: 10.2741/4852. Front Biosci (Landmark Ed). 2020. Retraction in: Front Biosci (Landmark Ed). 2023 Dec 29;28(12):369. doi: 10.31083/j.fbl2812369 PMID: 32114429 Retracted. Review.
-
Role for a Filamentous Nuclear Assembly of IFI16, DNA, and Host Factors in Restriction of Herpesviral Infection.mBio. 2019 Jan 22;10(1):e02621-18. doi: 10.1128/mBio.02621-18. mBio. 2019. PMID: 30670617 Free PMC article.
References
-
- Chemudupati M., Kenney A.D., Bonifati S., Zani A., McMichael T.M., Wu L., Yount J.S. From APOBEC to ZAP: Diverse mechanisms used by cellular restriction factors to inhibit virus infections. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2019;1866:382–394. doi: 10.1016/j.bbamcr.2018.09.012. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

