"Time Is out of Joint" in Pluripotent Stem Cells: How and Why
- PMID: 38396740
- PMCID: PMC10889767
- DOI: 10.3390/ijms25042063
"Time Is out of Joint" in Pluripotent Stem Cells: How and Why
Abstract
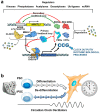
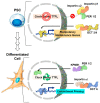
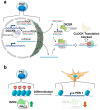
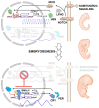
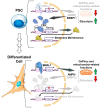
The circadian rhythm is necessary for the homeostasis and health of living organisms. Molecular clocks interconnected by transcription/translation feedback loops exist in most cells of the body. A puzzling exemption to this, otherwise, general biological hallmark is given by the cell physiology of pluripotent stem cells (PSCs) that lack circadian oscillations gradually acquired following their in vivo programmed differentiation. This process can be nicely phenocopied following in vitro commitment and reversed during the reprogramming of somatic cells to induce PSCs. The current understanding of how and why pluripotency is "time-uncoupled" is largely incomplete. A complex picture is emerging where the circadian core clockwork is negatively regulated in PSCs at the post-transcriptional/translational, epigenetic, and other-clock-interaction levels. Moreover, non-canonical functions of circadian core-work components in the balance between pluripotency identity and metabolic-driven cell reprogramming are emerging. This review selects and discusses results of relevant recent investigations providing major insights into this context.
Keywords: cellular differentiation; circadian rhythm; clock genes; pluripotent stem cells; reprogramming.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
The circadian clock CRY1 regulates pluripotent stem cell identity and somatic cell reprogramming.Cell Rep. 2023 Jun 27;42(6):112590. doi: 10.1016/j.celrep.2023.112590. Epub 2023 May 31. Cell Rep. 2023. PMID: 37261952
-
Development of the Circadian Core Machinery in Mammals.J Mol Biol. 2020 May 29;432(12):3611-3617. doi: 10.1016/j.jmb.2019.11.026. Epub 2020 Jan 10. J Mol Biol. 2020. PMID: 31931007 Review.
-
Role of circadian gene Clock during differentiation of mouse pluripotent stem cells.Protein Cell. 2016 Nov;7(11):820-832. doi: 10.1007/s13238-016-0319-9. Epub 2016 Sep 23. Protein Cell. 2016. PMID: 27664156 Free PMC article.
-
Pluripotency, Differentiation, and Reprogramming: A Gene Expression Dynamics Model with Epigenetic Feedback Regulation.PLoS Comput Biol. 2015 Aug 26;11(8):e1004476. doi: 10.1371/journal.pcbi.1004476. eCollection 2015 Aug. PLoS Comput Biol. 2015. PMID: 26308610 Free PMC article.
-
Molecular mechanisms of the circadian clockwork in mammals.FEBS Lett. 2014 Aug 1;588(15):2477-83. doi: 10.1016/j.febslet.2014.06.005. Epub 2014 Jun 6. FEBS Lett. 2014. PMID: 24911207 Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

