Influence of microbiota-associated metabolic reprogramming on clinical outcome in patients with melanoma from the randomized adjuvant dendritic cell-based MIND-DC trial
- PMID: 38395948
- PMCID: PMC10891084
- DOI: 10.1038/s41467-024-45357-1
Influence of microbiota-associated metabolic reprogramming on clinical outcome in patients with melanoma from the randomized adjuvant dendritic cell-based MIND-DC trial
Abstract
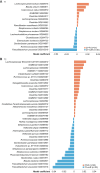
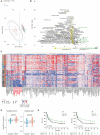
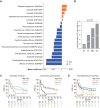
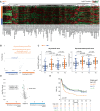
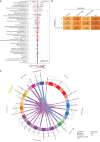
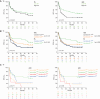
Tumor immunosurveillance plays a major role in melanoma, prompting the development of immunotherapy strategies. The gut microbiota composition, influencing peripheral and tumoral immune tonus, earned its credentials among predictors of survival in melanoma. The MIND-DC phase III trial (NCT02993315) randomized (2:1 ratio) 148 patients with stage IIIB/C melanoma to adjuvant treatment with autologous natural dendritic cell (nDC) or placebo (PL). Overall, 144 patients collected serum and stool samples before and after 2 bimonthly injections to perform metabolomics (MB) and metagenomics (MG) as prespecified exploratory analysis. Clinical outcomes are reported separately. Here we show that different microbes were associated with prognosis, with the health-related Faecalibacterium prausnitzii standing out as the main beneficial taxon for no recurrence at 2 years (p = 0.008 at baseline, nDC arm). Therapy coincided with major MB perturbations (acylcarnitines, carboxylic and fatty acids). Despite randomization, nDC arm exhibited MG and MB bias at baseline: relative under-representation of F. prausnitzii, and perturbations of primary biliary acids (BA). F. prausnitzii anticorrelated with BA, medium- and long-chain acylcarnitines. Combined, these MG and MB biomarkers markedly determined prognosis. Altogether, the host-microbial interaction may play a role in localized melanoma. We value systematic MG and MB profiling in randomized trials to avoid baseline differences attributed to host-microbe interactions.
© 2024. The Author(s).
Conflict of interest statement
L.Zi. founded EverImmune and is the SAB president of EverImmune. L.ZI. had grant support from Daichi Sankyo, Kaleido, 9 meters and Pileje. A.M.M.E.: consulting fees from Acetra, Agenus, BioInvent, Brenus, CatalYm, Epics, Ellipses, Galecto, GenOway, IO Biotech, IQVIA, ISA Pharmaceuticals, Merck&Co, MSD, Pierre Fabre, Sairopa, Scorpion, Sellas, SkylineDX, TigeTx, Trained Immunity TX. A.M.M.E.: participation on a Data Safety Monitoring Board: Boehringer Ingelheim, BioNTech, and Pfizer. A.M.M.E.: lectures for BMS, MSD. A.M.M.E.: stock or stock options for IO Biotech, SkylineDx and Sairopa. G.K. has been holding research contracts with Daiichi Sankyo, Eleor, Kaleido, Lytix Pharma, PharmaMar, Osasuna Therapeutics, Samsara Therapeutics, Sanofi, Tollys, and Vascage. G.K. is on the Board of Directors of the Bristol Myers Squibb Foundation France. G.K. is a scientific co-founder of EverImmune, Osasuna Therapeutics, Samsara Therapeutics and Therafast Bio. G.K. is in the scientific advisory boards of Hevolution, Institut Servier and Longevity Vision Funds. G.K. is the inventor of patents covering therapeutic targeting of aging, cancer, cystic fibrosis and metabolic disorders. G.K.’s brother, Romano Kroemer, was an employee of Sanofi and now consults for Boehringer-Ingelheim. B.R. is co-founder of Science Curebiota. L.D. is a SAB member of EverImmune. K.F.B.: consulting fees (institutional) from MSD and Pierre Fabre. The remaining authors declare no competing interests.
Figures
Similar articles
-
Adjuvant dendritic cell therapy in stage IIIB/C melanoma: the MIND-DC randomized phase III trial.Nat Commun. 2024 Feb 23;15(1):1632. doi: 10.1038/s41467-024-45358-0. Nat Commun. 2024. PMID: 38395969 Free PMC article. Clinical Trial.
-
Consistent Stool Metagenomic Biomarkers Associated with the Response To Melanoma Immunotherapy.mSystems. 2023 Apr 27;8(2):e0102322. doi: 10.1128/msystems.01023-22. Epub 2023 Feb 21. mSystems. 2023. PMID: 36809182 Free PMC article.
-
Prospective, randomized, double-blind phase 2B trial of the TLPO and TLPLDC vaccines to prevent recurrence of resected stage III/IV melanoma: a prespecified 36-month analysis.J Immunother Cancer. 2023 Aug;11(8):e006665. doi: 10.1136/jitc-2023-006665. J Immunother Cancer. 2023. PMID: 37536936 Free PMC article. Clinical Trial.
-
The role of adjuvant therapy in melanoma management.Cancer. 1995 Jan 15;75(2 Suppl):726-34. doi: 10.1002/1097-0142(19950115)75:2+<726::aid-cncr2820751417>3.0.co;2-r. Cancer. 1995. PMID: 7805001 Review.
-
Neoadjuvant treatment for stage III and IV cutaneous melanoma.Cochrane Database Syst Rev. 2023 Jan 17;1(1):CD012974. doi: 10.1002/14651858.CD012974.pub2. Cochrane Database Syst Rev. 2023. PMID: 36648215 Free PMC article. Review.
Cited by
-
Immune Checkpoint Inhibitor Therapy for Metastatic Melanoma: What Should We Focus on to Improve the Clinical Outcomes?Int J Mol Sci. 2024 Sep 20;25(18):10120. doi: 10.3390/ijms251810120. Int J Mol Sci. 2024. PMID: 39337605 Free PMC article. Review.
-
Faecalibaterium prausnitzii strain EXL01 boosts efficacy of immune checkpoint inhibitors.Oncoimmunology. 2024 Jul 1;13(1):2374954. doi: 10.1080/2162402X.2024.2374954. eCollection 2024. Oncoimmunology. 2024. PMID: 38957477 Free PMC article.
-
ACBP/DBI neutralization for the experimental treatment of fatty liver disease.Cell Death Differ. 2024 Nov 16. doi: 10.1038/s41418-024-01410-6. Online ahead of print. Cell Death Differ. 2024. PMID: 39550516
-
Disentangling the molecular mystery of tumour-microbiota interactions: Microbial metabolites.Clin Transl Med. 2024 Nov;14(11):e70093. doi: 10.1002/ctm2.70093. Clin Transl Med. 2024. PMID: 39568157 Free PMC article. Review.
-
Adjuvant dendritic cell therapy in stage IIIB/C melanoma: the MIND-DC randomized phase III trial.Nat Commun. 2024 Feb 23;15(1):1632. doi: 10.1038/s41467-024-45358-0. Nat Commun. 2024. PMID: 38395969 Free PMC article. Clinical Trial.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

