An Expeditious Neutralization Assay for Porcine Reproductive and Respiratory Syndrome Virus Based on a Recombinant Virus Expressing Green Fluorescent Protein
- PMID: 38392184
- PMCID: PMC10887926
- DOI: 10.3390/cimb46020066
An Expeditious Neutralization Assay for Porcine Reproductive and Respiratory Syndrome Virus Based on a Recombinant Virus Expressing Green Fluorescent Protein
Abstract
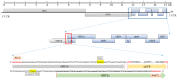
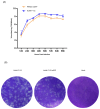
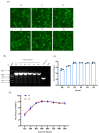
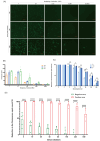
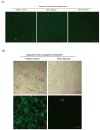
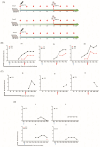
Due to the extensive genetic and antigenic variation in Porcine Reproductive and Respiratory Syndrome Virus (PRRSV), as well as its rapid mutability and evolution, PRRS prevention and control can be challenging. An expeditious and sensitive neutralization assay for PRRSV is presented to monitor neutralizing antibodies (NAbs) in serum during vaccine research. Here, a PRRSV expressing eGFP was successfully rescued with reverse genetics based on the infectious clone HuN4-F112-eGFP which we constructed. The fluorescent protein expressions of the reporter viruses remained stable for at least five passages. Based on this reporter virus, the neutralization assay can be easily used to evaluate the level of NAbs by counting cells with green fluorescence. Compared with the classical CPE assay, the newly developed assay increases sensitivity by one- to four-fold at the early antibody response stage, thus saving 2 days of assay waiting time. By using this assay to unveil the dynamics of neutralizing antibodies against PRRSV, priming immunity through either a single virulent challenge or only vaccination could produce limited NAbs, but re-infection with PRRSV would induce a faster and stronger NAb response. Overall, the novel HuN4-F112-eGFP-based neutralization assay holds the potential to provide a highly efficient platform for evaluating the next generation of PRRS vaccines.
Keywords: PRRSV; enhanced GFP; infectious clones; neutralization assay; neutralizing antibodies; reporter virus.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
ORF1a of highly pathogenic PRRS attenuated vaccine virus plays a key role in neutralizing antibody induction in piglets and virus neutralization in vitro.Virol J. 2017 Aug 22;14(1):159. doi: 10.1186/s12985-017-0825-2. Virol J. 2017. PMID: 28830563 Free PMC article.
-
Clover-tagged porcine reproductive and respiratory syndrome virus infectious clones for rapid detection of virus neutralizing antibodies.J Virol Methods. 2018 Sep;259:100-105. doi: 10.1016/j.jviromet.2018.06.013. Epub 2018 Jun 24. J Virol Methods. 2018. PMID: 29949736
-
[Study on using NSP2 protein of porcine reproductive and respiratory syndrome virus (HuN4-F112) to express E2 neutralizing epitope of classical swine fever virus].Bing Du Xue Bao. 2013 Jan;29(1):17-25. Bing Du Xue Bao. 2013. PMID: 23547375 Chinese.
-
Innate and adaptive immunity against Porcine Reproductive and Respiratory Syndrome Virus.Vet Immunol Immunopathol. 2015 Sep 15;167(1-2):1-14. doi: 10.1016/j.vetimm.2015.07.003. Epub 2015 Jul 17. Vet Immunol Immunopathol. 2015. PMID: 26209116 Free PMC article. Review.
-
Porcine reproductive and respiratory syndrome virus vaccines: current status and strategies to a universal vaccine.Transbound Emerg Dis. 2014 Apr;61(2):109-20. doi: 10.1111/tbed.12016. Epub 2013 Jan 24. Transbound Emerg Dis. 2014. PMID: 23343057 Review.
References
-
- Neumann E.J., Kliebenstein J.B., Johnson C.D., Mabry J.W., Bush E.J., Seitzinger A.H., Green A.L., Zimmerman J.J. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J. Am. Vet. Med. Assoc. 2005;227:385–392. doi: 10.2460/javma.2005.227.385. - DOI - PubMed
-
- Mengeling W.L., Lager K.M., Vorwald A.C. Clinical consequences of exposing pregnant gilts to strains of porcine reproductive and respiratory syndrome (PRRS) virus isolated from field cases of “atypical” PRRS. Am. J. Vet. Res. 1998;59:1540–1544. - PubMed
-
- Allende R., Lewis T.L., Lu Z., Rock D.L., Kutish G.F., Ali A., Doster A.R., Osorio F.A. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. Pt 2J. Gen. Virol. 1999;80:307–315. doi: 10.1099/0022-1317-80-2-307. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

