The Long Non-Coding RNA NR3C2-8:1 Promotes p53-Mediated Apoptosis through the miR-129-5p/USP10 Axis in Amyotrophic Lateral Sclerosis
- PMID: 38388775
- PMCID: PMC11415439
- DOI: 10.1007/s12035-024-04059-x
The Long Non-Coding RNA NR3C2-8:1 Promotes p53-Mediated Apoptosis through the miR-129-5p/USP10 Axis in Amyotrophic Lateral Sclerosis
Abstract
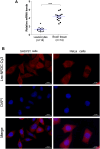
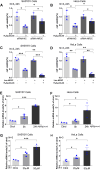
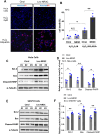
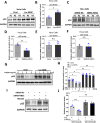
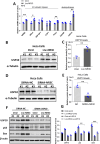
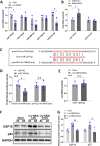
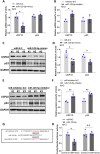
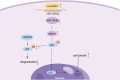
Motor neuron degeneration in amyotrophic lateral sclerosis (ALS) is a form of apoptosis, but the mechanisms underlying this neuronal cell death remain unclear. Numerous studies demonstrate abnormally elevated and active p53 in the central nervous system of ALS patients. Activation of p53-regulated pro-apoptotic signaling pathways may trigger motor neuron death. We previously reported decreased expression of the long non-coding RNA NR3C2-8:1 (Lnc-NR3C) in leukocytes of ALS patients. Here, we show lnc-NR3C promotes p53-mediated cell death in ALS by upregulating USP10 and promoting lnc-NR3C-triggered p53 activation, resulting in cell death. Conversely, lnc-NR3C knockdown inhibited USP10-triggered p53 activation, thereby protecting cells against oxidative stress. As a competitive endogenous RNA, lnc-NR3C competitively binds miR-129-5p, regulating the usp10/p53 axis. Elucidating the link between Lnc-NR3C and the USP10/p53 axis in an ALS cell model reveals a role for long non-coding RNAs in activating apoptosis. This provides new therapeutic opportunities in ALS.
© 2024. The Author(s).
Conflict of interest statement
The authors confirm that there are no conflicts of interest.
Figures
Similar articles
-
Lnc-HIBADH-4 Regulates Autophagy-Lysosome Pathway in Amyotrophic Lateral Sclerosis by Targeting Cathepsin D.Mol Neurobiol. 2024 Jul;61(7):4768-4782. doi: 10.1007/s12035-023-03835-5. Epub 2023 Dec 22. Mol Neurobiol. 2024. PMID: 38135852 Free PMC article.
-
Downregulation of Lnc-ABCA12-3 modulates UBQLN1 expression and protein homeostasis pathways in amyotrophic lateral sclerosis.Sci Rep. 2024 Sep 13;14(1):21383. doi: 10.1038/s41598-024-72666-8. Sci Rep. 2024. PMID: 39271939 Free PMC article.
-
Hypoxia-induced long non-coding RNA LINC00460 promotes p53 mediated proliferation and metastasis of pancreatic cancer by regulating the miR-4689/UBE2V1 axis and sequestering USP10.Int J Med Sci. 2023 Sep 4;20(10):1339-1357. doi: 10.7150/ijms.87833. eCollection 2023. Int J Med Sci. 2023. PMID: 37786443 Free PMC article.
-
MicroRNAs as regulators of cell death mechanisms in amyotrophic lateral sclerosis.J Cell Mol Med. 2019 Mar;23(3):1647-1656. doi: 10.1111/jcmm.13976. Epub 2019 Jan 4. J Cell Mol Med. 2019. PMID: 30614179 Free PMC article. Review.
-
MicroRNA Metabolism and Dysregulation in Amyotrophic Lateral Sclerosis.Mol Neurobiol. 2018 Mar;55(3):2617-2630. doi: 10.1007/s12035-017-0537-z. Epub 2017 Apr 18. Mol Neurobiol. 2018. PMID: 28421535 Review.
Cited by
-
Expression Changes of miRNAs in Humans and Animal Models of Amyotrophic Lateral Sclerosis and Their Potential Application for Clinical Diagnosis.Life (Basel). 2024 Sep 6;14(9):1125. doi: 10.3390/life14091125. Life (Basel). 2024. PMID: 39337908 Free PMC article. Review.
References
-
- Sathasivam S, Shaw PJ (2005) Apoptosis in amyotrophic lateral sclerosis–what is the evidence? Lancet Neurol 4(8):500–509 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

