Immune mapping of human tuberculosis and sarcoidosis lung granulomas
- PMID: 38385142
- PMCID: PMC10879604
- DOI: 10.3389/fimmu.2023.1332733
Immune mapping of human tuberculosis and sarcoidosis lung granulomas
Abstract
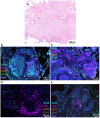
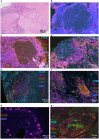
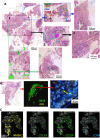
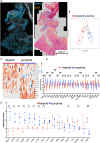
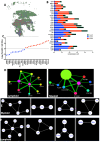
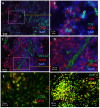
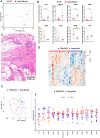
Tuberculosis (TB) and sarcoidosis are both granulomatous diseases. Here, we compared the immunological microenvironments of granulomas from TB and sarcoidosis patients using in situ sequencing (ISS) transcriptomic analysis and multiplexed immunolabeling of tissue sections. TB lesions consisted of large necrotic and cellular granulomas, whereas "multifocal" granulomas with macrophages or epitheloid cell core and a T-cell rim were observed in sarcoidosis samples. The necrotic core in TB lesions was surrounded by macrophages and encircled by a dense T-cell layer. Within the T-cell layer, compact B-cell aggregates were observed in most TB samples. These B-cell clusters were vascularized and could contain defined B-/T-cell and macrophage-rich areas. The ISS of 40-60 immune transcripts revealed the enriched expression of transcripts involved in homing or migration to lymph nodes, which formed networks at single-cell distances in lymphoid areas of the TB lesions. Instead, myeloid-annotated regions were enriched in CD68, CD14, ITGAM, ITGAX, and CD4 mRNA. CXCL8 and IL1B mRNA were observed in granulocytic areas in which M. tuberculosis was also detected. In line with ISS data indicating tertiary lymphoid structures, immune labeling of TB sections expressed markers of high endothelial venules, follicular dendritic cells, follicular helper T cells, and lymph-node homing receptors on T cells. Neither ISS nor immunolabeling showed evidence of tertiary lymphoid aggregates in sarcoidosis samples. Together, our finding suggests that despite their heterogeneity, the formation of tertiary immune structures is a common feature in granulomas from TB patients.
Keywords: granuloma; inducible bronchus associated lymphoid tissue; lung; sarcoidosis; spatial transcriptomics; tuberculosis.
Copyright © 2024 Carow, Muliadi, Skålén, Yokota, Kathamuthu, Setiabudiawan, Lange, Scheu, Gaede, Goldmann, Pandita, Masood, Pervez, Grunewald, Hasan, Levin and Rottenberg.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
Similar articles
-
Evidence for M2 macrophages in granulomas from pulmonary sarcoidosis: A new aspect of macrophage heterogeneity.Hum Immunol. 2018 Jan;79(1):63-69. doi: 10.1016/j.humimm.2017.10.009. Epub 2017 Oct 31. Hum Immunol. 2018. PMID: 29107084
-
Diverse immune environments in human lung tuberculosis granulomas assessed by quantitative multiplexed immunofluorescence.Mod Pathol. 2020 Dec;33(12):2507-2519. doi: 10.1038/s41379-020-0600-6. Epub 2020 Jun 26. Mod Pathol. 2020. PMID: 32591586
-
Spatial multiomic profiling reveals the novel polarization of foamy macrophages within necrotic granulomatous lesions developed in lungs of C3HeB/FeJ mice infected with Mycobacterium tuberculosis.Front Cell Infect Microbiol. 2022 Sep 27;12:968543. doi: 10.3389/fcimb.2022.968543. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36237431 Free PMC article.
-
The pathology of pulmonary sarcoidosis: update.Semin Diagn Pathol. 2007 Aug;24(3):150-61. doi: 10.1053/j.semdp.2007.06.002. Semin Diagn Pathol. 2007. PMID: 17882899 Review.
-
Reinventing the human tuberculosis (TB) granuloma: Learning from the cancer field.Front Immunol. 2022 Dec 15;13:1059725. doi: 10.3389/fimmu.2022.1059725. eCollection 2022. Front Immunol. 2022. PMID: 36591229 Free PMC article. Review.
References
-
- WHO . Global tuberculosis report 2022. Geneva: World Health Organization; (2022).
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

