Antibody profiling and predictive modeling discriminate between Kaposi sarcoma and asymptomatic KSHV infection
- PMID: 38381773
- PMCID: PMC10911871
- DOI: 10.1371/journal.ppat.1012023
Antibody profiling and predictive modeling discriminate between Kaposi sarcoma and asymptomatic KSHV infection
Abstract
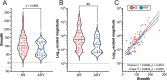
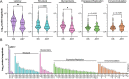
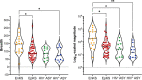
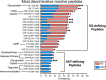
Protein-level immunodominance patterns against Kaposi sarcoma-associated herpesvirus (KSHV), the aetiologic agent of Kaposi sarcoma (KS), have been revealed from serological probing of whole protein arrays, however, the epitopes that underlie these patterns have not been defined. We recently demonstrated the utility of phage display in high-resolution linear epitope mapping of the KSHV latency-associated nuclear antigen (LANA/ORF73). Here, a VirScan phage immunoprecipitation and sequencing approach, employing a library of 1,988 KSHV proteome-derived peptides, was used to quantify the breadth and magnitude of responses of 59 sub-Saharan African KS patients and 22 KSHV-infected asymptomatic individuals (ASY), and ultimately to support an application of machine-learning-based predictive modeling using the peptide-level responses. Comparing anti-KSHV antibody repertoire revealed that magnitude, not breadth, increased in KS. The most targeted epitopes in both KS and ASY were in the immunodominant proteins, notably, K8.129-56 and ORF65140-168, in addition to LANA. Finally, using unbiased machine-learning-based predictive models, reactivity to a subset of 25 discriminative peptides was demonstrated to successfully classify KS patients from asymptomatic individuals. Our study provides the highest resolution mapping of antigenicity across the entire KSHV proteome to date, which is vital to discern mechanisms of viral pathogenesis, to define prognostic biomarkers, and to design effective vaccine and therapeutic strategies. Future studies will investigate the diagnostic, prognostic, and therapeutic potential of the 25 discriminative peptides.
Copyright: © 2024 Bennett et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Antibody epitope profiling of the KSHV LANA protein using VirScan.PLoS Pathog. 2022 Dec 19;18(12):e1011033. doi: 10.1371/journal.ppat.1011033. eCollection 2022 Dec. PLoS Pathog. 2022. PMID: 36534707 Free PMC article.
-
Differences in Kaposi sarcoma-associated herpesvirus-specific and herpesvirus-non-specific immune responses in classic Kaposi sarcoma cases and matched controls in Sicily.Cancer Sci. 2011 Oct;102(10):1769-73. doi: 10.1111/j.1349-7006.2011.02032.x. Epub 2011 Jul 28. Cancer Sci. 2011. PMID: 21740480 Free PMC article.
-
Activated Nrf2 Interacts with Kaposi's Sarcoma-Associated Herpesvirus Latency Protein LANA-1 and Host Protein KAP1 To Mediate Global Lytic Gene Repression.J Virol. 2015 Aug;89(15):7874-92. doi: 10.1128/JVI.00895-15. Epub 2015 May 20. J Virol. 2015. PMID: 25995248 Free PMC article.
-
Viral latent proteins as targets for Kaposi's sarcoma and Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) induced lymphoma.Curr Drug Targets Infect Disord. 2003 Jun;3(2):129-35. doi: 10.2174/1568005033481150. Curr Drug Targets Infect Disord. 2003. PMID: 12769790 Review.
-
Diagnosis and Treatment of Kaposi Sarcoma.Am J Clin Dermatol. 2017 Aug;18(4):529-539. doi: 10.1007/s40257-017-0270-4. Am J Clin Dermatol. 2017. PMID: 28324233 Free PMC article. Review.
Cited by
-
Comprehensive phage display viral antibody profiling using VirScan: potential applications in chronic immune-mediated disease.J Virol. 2024 Nov 19;98(11):e0110224. doi: 10.1128/jvi.01102-24. Epub 2024 Oct 21. J Virol. 2024. PMID: 39431820 Review.
References
-
- Butler LM, Dorsey G, Hladik W, Rosenthal PJ, Brander C, Neilands TB, et al.. Kaposi Sarcoma–Associated Herpesvirus (KSHV) Seroprevalence in Population-Based Samples of African Children: Evidence for At Least 2 Patterns of KSHV Transmission. J Infect Dis. 2009;200: 430–438. doi: 10.1086/600103 - DOI - PMC - PubMed
-
- Minhas V, Crabtree KL, Chao A, M’soka TJ, Kankasa C, Bulterys M, et al.. Early Childhood Infection by Human Herpesvirus 8 in Zambia and the Role of Human Immunodeficiency Virus Type 1 Coinfection in a Highly Endemic Area. Am J Epidemiol. 2008;168: 311–320. doi: 10.1093/aje/kwn125 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

