Long-read sequencing reveals the structural complexity of genomic integration of HPV DNA in cervical cancer cell lines
- PMID: 38378450
- PMCID: PMC10877919
- DOI: 10.1186/s12864-024-10101-y
Long-read sequencing reveals the structural complexity of genomic integration of HPV DNA in cervical cancer cell lines
Abstract
Background: Cervical cancer (CC) causes more than 311,000 deaths annually worldwide. The integration of human papillomavirus (HPV) is a crucial genetic event that contributes to cervical carcinogenesis. Despite HPV DNA integration is known to disrupt the genomic architecture of both the host and viral genomes in CC, the complexity of this process remains largely unexplored.
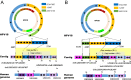
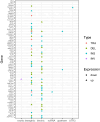
Results: In this study, we conducted whole-genome sequencing (WGS) at 55-65X coverage utilizing the PacBio long-read sequencing platform in SiHa and HeLa cells, followed by comprehensive analyses of the sequence data to elucidate the complexity of HPV integration. Firstly, our results demonstrated that PacBio long-read sequencing effectively identifies HPV integration breakpoints with comparable accuracy to targeted-capture Next-generation sequencing (NGS) methods. Secondly, we constructed detailed models of complex integrated genome structures that included both the HPV genome and nearby regions of the human genome by utilizing PacBio long-read WGS. Thirdly, our sequencing results revealed the occurrence of a wide variety of genome-wide structural variations (SVs) in SiHa and HeLa cells. Additionally, our analysis further revealed a potential correlation between changes in gene expression levels and SVs on chromosome 13 in the genome of SiHa cells.
Conclusions: Using PacBio long-read sequencing, we have successfully constructed complex models illustrating HPV integrated genome structures in SiHa and HeLa cells. This accomplishment serves as a compelling demonstration of the valuable capabilities of long-read sequencing in detecting and characterizing HPV genomic integration structures within human cells. Furthermore, these findings offer critical insights into the complex process of HPV16 and HPV18 integration and their potential contribution to the development of cervical cancer.
Keywords: Cervical cancer; HPV integration; HPV16; HPV18.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Comprehensive mapping of the human papillomavirus (HPV) DNA integration sites in cervical carcinomas by HPV capture technology.Oncotarget. 2016 Feb 2;7(5):5852-64. doi: 10.18632/oncotarget.6809. Oncotarget. 2016. PMID: 26735580 Free PMC article.
-
Integrated HPV genomes tend to integrate in gene desert areas in the CaSki, HeLa, and SiHa cervical cancer cell lines.Life Sci. 2015 Apr 15;127:46-52. doi: 10.1016/j.lfs.2015.01.039. Epub 2015 Mar 5. Life Sci. 2015. PMID: 25747255
-
Long-read sequencing unveils high-resolution HPV integration and its oncogenic progression in cervical cancer.Nat Commun. 2022 May 10;13(1):2563. doi: 10.1038/s41467-022-30190-1. Nat Commun. 2022. PMID: 35538075 Free PMC article.
-
Human papillomavirus genome integration in squamous carcinogenesis: what have next-generation sequencing studies taught us?J Pathol. 2018 May;245(1):9-18. doi: 10.1002/path.5058. Epub 2018 Mar 30. J Pathol. 2018. PMID: 29443391 Review.
-
Identification of HPV Integration and Genomic Patterns Delineating the Clinical Landscape of Cervical Cancer.Asian Pac J Cancer Prev. 2015;16(18):8041-5. doi: 10.7314/apjcp.2015.16.18.8041. Asian Pac J Cancer Prev. 2015. PMID: 26745036 Review.
Cited by
-
Identification of novel genomic hotspots and tumor-relevant genes via comprehensive analysis of HPV integration in Chinese patients of cervical cancer.Am J Cancer Res. 2024 Sep 25;14(9):4665-4682. doi: 10.62347/KKLE8602. eCollection 2024. Am J Cancer Res. 2024. PMID: 39417198 Free PMC article.
References
-
- Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189(1):12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. - DOI - PubMed
-
- Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995;87(11):796–802. doi: 10.1093/jnci/87.11.796. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

