Metagenomic survey of viral diversity obtained from feces of piglets with diarrhea
- PMID: 38375275
- PMCID: PMC10875384
- DOI: 10.1016/j.heliyon.2024.e25616
Metagenomic survey of viral diversity obtained from feces of piglets with diarrhea
Abstract
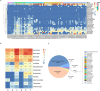
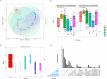
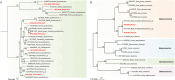
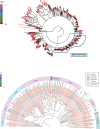
Pigs are natural host to various zoonotic pathogens including viruses. In this study, we analyzed the viral communities in the feces of 89 piglets with diarrhea under one month old which were collected from six farms in Jiangsu Province of the Eastern China, using the unbiased virus metagenomic method. A total of 89 libraries were constructed, and 46937894 unique sequence reads were generated by Illumina sequencing. Overall, the family Picornaviridae accounted for the majority of the total reads of putative mammalian viruses. Ten novel virus genomes from different family members were discovered, including Parvoviridae (n = 2), Picobirnaviridae (n = 4) and CRESS DNA viruses (n = 4). A large number of phages were identified, which mainly belonged to the order Caudovirales and the family Microviridae. Moreover, some identified viruses were closely related to viruses found in non-porcine hosts, highlighting the potential for cross-species virus dissemination. This study increased our understanding of the fecal virus communities of diarrhea piglets and provided valuable information for virus monitoring and preventing.
Keywords: Diarrhea piglets; Feces; Metagenomic; Viral communities; Virus genome.
© 2024 The Authors. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
The fecal virome of pigs on a high-density farm.J Virol. 2011 Nov;85(22):11697-708. doi: 10.1128/JVI.05217-11. Epub 2011 Sep 7. J Virol. 2011. PMID: 21900163 Free PMC article.
-
Case-Control Comparison of Enteric Viromes in Captive Rhesus Macaques with Acute or Idiopathic Chronic Diarrhea.J Virol. 2017 Aug 24;91(18):e00952-17. doi: 10.1128/JVI.00952-17. Print 2017 Sep 15. J Virol. 2017. PMID: 28659484 Free PMC article.
-
Viral metagenomics reveals diverse viruses in the fecal samples of children with diarrhea.Virol Sin. 2022 Feb;37(1):82-93. doi: 10.1016/j.virs.2022.01.012. Epub 2022 Jan 17. Virol Sin. 2022. PMID: 35234620 Free PMC article.
-
Viral Metagenomic Analysis of the Fecal Samples in Domestic Dogs (Canis lupus familiaris).Viruses. 2023 Mar 6;15(3):685. doi: 10.3390/v15030685. Viruses. 2023. PMID: 36992396 Free PMC article.
-
Comparison of viral communities in the blood, feces and various tissues of wild brown rats (Rattus norvegicus).Heliyon. 2023 Jun 13;9(6):e17222. doi: 10.1016/j.heliyon.2023.e17222. eCollection 2023 Jun. Heliyon. 2023. PMID: 37389044 Free PMC article.
References
-
- Qin P., Li H., Wang J.-W., et al. Genetic and pathogenic characterization of a novel reassortant mammalian orthoreovirus 3 (MRV3) from a diarrheic piglet and seroepidemiological survey of MRV3 in diarrheic pigs from east China [J] Vet. Microbiol. 2017;208:126–136. doi: 10.1016/j.vetmic.2017.07.021. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

