This is a preprint.
Transcription Factor Condensates Mediate Clustering of MET Regulon and Enhancement in Gene Expression
- PMID: 38370634
- PMCID: PMC10871269
- DOI: 10.1101/2024.02.06.579062
Transcription Factor Condensates Mediate Clustering of MET Regulon and Enhancement in Gene Expression
Update in
-
Transcription factor condensates, 3D clustering, and gene expression enhancement of the MET regulon.Elife. 2024 Sep 30;13:RP96028. doi: 10.7554/eLife.96028. Elife. 2024. PMID: 39347738 Free PMC article.
Abstract
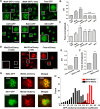
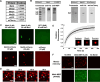
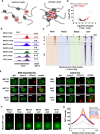
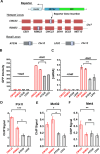
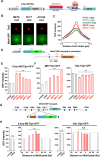
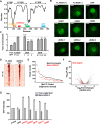
Some transcription factors (TFs) can form liquid-liquid phase separated (LLPS) condensates. However, the functions of these TF condensates in 3D genome organization and gene regulation remain elusive. In response to methionine (met) starvation, budding yeast TF Met4 and a few co-activators, including Met32, induce a set of genes involved in met biosynthesis. Here, we show that the endogenous Met4 and Met32 form co-localized puncta-like structures in yeast nuclei upon met depletion. Recombinant Met4 and Met32 form mixed droplets with LLPS properties in vitro. In relation to chromatin, Met4 puncta co-localize with target genes, and at least a subset of these target genes is clustered in 3D in a Met4-dependent manner. A MET3pr-GFP reporter inserted near several native Met4 binding sites becomes co-localized with Met4 puncta and displays enhanced transcriptional activity. A Met4 variant with a partial truncation of an intrinsically disordered region (IDR) shows less puncta formation, and this mutant selectively reduces the reporter activity near Met4 binding sites to the basal level. Overall, these results support a model where Met4 and co-activators form condensates to bring multiple target genes into a vicinity with higher local TF concentrations, which facilitates a strong response to methionine depletion.
Conflict of interest statement
Conflict of Interest Statement The authors declare no conflict of interest.
Figures

Similar articles
-
Transcription factor condensates, 3D clustering, and gene expression enhancement of the MET regulon.Elife. 2024 Sep 30;13:RP96028. doi: 10.7554/eLife.96028. Elife. 2024. PMID: 39347738 Free PMC article.
-
Characterizing the roles of Met31 and Met32 in coordinating Met4-activated transcription in the absence of Met30.Mol Biol Cell. 2012 May;23(10):1928-42. doi: 10.1091/mbc.E11-06-0532. Epub 2012 Mar 21. Mol Biol Cell. 2012. PMID: 22438580 Free PMC article.
-
A dominant suppressor mutation of the met30 cell cycle defect suggests regulation of the Saccharomyces cerevisiae Met4-Cbf1 transcription complex by Met32.J Biol Chem. 2008 Apr 25;283(17):11615-24. doi: 10.1074/jbc.M708230200. Epub 2008 Feb 28. J Biol Chem. 2008. PMID: 18308733 Free PMC article.
-
Super-Enhancers, Phase-Separated Condensates, and 3D Genome Organization in Cancer.Cancers (Basel). 2022 Jun 10;14(12):2866. doi: 10.3390/cancers14122866. Cancers (Basel). 2022. PMID: 35740532 Free PMC article. Review.
-
Unravelling the microscopic characteristics of intrinsically disordered proteins upon liquid-liquid phase separation.Essays Biochem. 2022 Dec 16;66(7):891-900. doi: 10.1042/EBC20220148. Essays Biochem. 2022. PMID: 36524527 Review.
References
-
- Apostolou E, Thanos D. 2008. Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell 134: 85–96. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
