This is a preprint.
Sparse CBX2 nucleates many Polycomb proteins to promote facultative heterochromatinization of Polycomb target genes
- PMID: 38370615
- PMCID: PMC10871256
- DOI: 10.1101/2024.02.05.578969
Sparse CBX2 nucleates many Polycomb proteins to promote facultative heterochromatinization of Polycomb target genes
Abstract
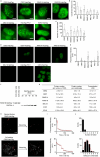
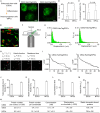
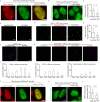
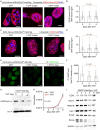
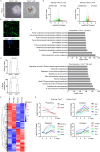
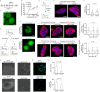
Facultative heterochromatinization of genomic regulators by Polycomb repressive complex (PRC) 1 and 2 is essential in development and differentiation; however, the underlying molecular mechanisms remain obscure. Using genetic engineering, molecular approaches, and live-cell single-molecule imaging, we quantify the number of proteins within condensates formed through liquid-liquid phase separation (LLPS) and find that in mouse embryonic stem cells (mESCs), approximately 3 CBX2 proteins nucleate many PRC1 and PRC2 subunits to form one non-stoichiometric condensate. We demonstrate that sparse CBX2 prevents Polycomb proteins from migrating to constitutive heterochromatin, demarcates the spatial boundaries of facultative heterochromatin, controls the deposition of H3K27me3, regulates transcription, and impacts cellular differentiation. Furthermore, we show that LLPS of CBX2 is required for the demarcation and deposition of H3K27me3 and is essential for cellular differentiation. Our findings uncover new functional roles of LLPS in the formation of facultative heterochromatin and unravel a new mechanism by which low-abundant proteins nucleate many other proteins to form compartments that enable them to execute their functions.
Keywords: CBX2; H3K27me3; PRC1; PRC2; Polycomb; chromatin; constitutive heterochromatin; facultative heterochromatin; liquid-liquid phase separation; live-cell single-molecule tracking; nucleation; single-molecule imaging.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing financial interest.
Figures

Similar articles
-
Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation.J Biol Chem. 2019 Feb 1;294(5):1451-1463. doi: 10.1074/jbc.RA118.006620. Epub 2018 Dec 4. J Biol Chem. 2019. PMID: 30514760 Free PMC article.
-
Phase-Separated Transcriptional Condensates Accelerate Target-Search Process Revealed by Live-Cell Single-Molecule Imaging.Cell Rep. 2020 Oct 13;33(2):108248. doi: 10.1016/j.celrep.2020.108248. Cell Rep. 2020. PMID: 33053359 Free PMC article.
-
PRC2.1- and PRC2.2-specific accessory proteins drive recruitment of different forms of canonical PRC1.Mol Cell. 2023 May 4;83(9):1393-1411.e7. doi: 10.1016/j.molcel.2023.03.018. Epub 2023 Apr 7. Mol Cell. 2023. PMID: 37030288 Free PMC article.
-
Beyond EZH2: is the polycomb protein CBX2 an emerging target for anti-cancer therapy?Expert Opin Ther Targets. 2019 Jul;23(7):565-578. doi: 10.1080/14728222.2019.1627329. Epub 2019 Jun 10. Expert Opin Ther Targets. 2019. PMID: 31177918 Review.
-
Biological functions of chromobox (CBX) proteins in stem cell self-renewal, lineage-commitment, cancer and development.Bone. 2021 Feb;143:115659. doi: 10.1016/j.bone.2020.115659. Epub 2020 Sep 24. Bone. 2021. PMID: 32979540 Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
