Lysine-36 of Drosophila histone H3.3 supports adult longevity
- PMID: 38366796
- PMCID: PMC10989886
- DOI: 10.1093/g3journal/jkae030
Lysine-36 of Drosophila histone H3.3 supports adult longevity
Abstract
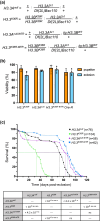
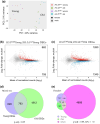
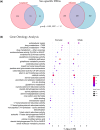
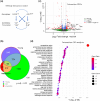
Aging is a multifactorial process that disturbs homeostasis, increases disease susceptibility, and ultimately results in death. Although the definitive set of molecular mechanisms responsible for aging remain to be discovered, epigenetic change over time is proving to be a promising piece of the puzzle. Several post-translational histone modifications have been linked to the maintenance of longevity. Here, we focus on lysine-36 of the replication-independent histone protein, H3.3 (H3.3K36). To interrogate the role of this residue in Drosophila developmental gene regulation, we generated a lysine-to-arginine mutant that blocks the activity of its cognate-modifying enzymes. We found that an H3.3BK36R mutation causes a significant reduction in adult lifespan, accompanied by dysregulation of the genomic and transcriptomic architecture. Transgenic co-expression of wild-type H3.3B completely rescues the longevity defect. Because H3.3 is known to accumulate in nondividing tissues, we carried out transcriptome profiling of young vs aged adult fly heads. The data show that loss of H3.3K36 results in age-dependent misexpression of NF-κB and other innate immune target genes, as well as defects in silencing of heterochromatin. We propose H3.3K36 maintains the postmitotic epigenomic landscape, supporting longevity by regulating both pericentric and telomeric retrotransposons and by suppressing aberrant immune signaling.
Keywords: PTMs; aging; gene expression; heterochromatin; histone variant; innate immunity; post-transcriptional modification; retrotransposition; senescence; telomeres.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest. The author(s) declare no conflict of interest.
Figures

Update of
-
Lysine-36 of Drosophila histone H3.3 supports adult longevity.bioRxiv [Preprint]. 2023 Dec 13:2023.09.28.559962. doi: 10.1101/2023.09.28.559962. bioRxiv. 2023. Update in: G3 (Bethesda). 2024 Apr 3;14(4):jkae030. doi: 10.1093/g3journal/jkae030 PMID: 38196611 Free PMC article. Updated. Preprint.
Similar articles
-
Lysine-36 of Drosophila histone H3.3 supports adult longevity.bioRxiv [Preprint]. 2023 Dec 13:2023.09.28.559962. doi: 10.1101/2023.09.28.559962. bioRxiv. 2023. Update in: G3 (Bethesda). 2024 Apr 3;14(4):jkae030. doi: 10.1093/g3journal/jkae030 PMID: 38196611 Free PMC article. Updated. Preprint.
-
Functional Redundancy of Variant and Canonical Histone H3 Lysine 9 Modification in Drosophila.Genetics. 2018 Jan;208(1):229-244. doi: 10.1534/genetics.117.300480. Epub 2017 Nov 13. Genetics. 2018. PMID: 29133298 Free PMC article.
-
Molecular landscape of modified histones in Drosophila heterochromatic genes and euchromatin-heterochromatin transition zones.PLoS Genet. 2008 Jan;4(1):e16. doi: 10.1371/journal.pgen.0040016. Epub 2007 Dec 13. PLoS Genet. 2008. PMID: 18208336 Free PMC article.
-
The indexing potential of histone lysine methylation.Novartis Found Symp. 2004;259:22-37; discussion 37-47, 163-9. Novartis Found Symp. 2004. PMID: 15171245 Review.
-
[Epigenetic regulation of histone H3 lysine 9 methylation in leukemia].Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2012 Feb;20(1):210-3. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2012. PMID: 22391199 Review. Chinese.
Cited by
-
Set2 and H3K36 regulate the Drosophila male X chromosome in a context-specific manner, independent from MSL complex spreading.bioRxiv [Preprint]. 2024 May 6:2024.05.03.592390. doi: 10.1101/2024.05.03.592390. bioRxiv. 2024. Update in: Genetics. 2024 Oct 17:iyae168. doi: 10.1093/genetics/iyae168 PMID: 38766267 Free PMC article. Updated. Preprint.
-
Genomic context-dependent histone H3K36 methylation by three Drosophila methyltransferases and implications for dedicated chromatin readers.Nucleic Acids Res. 2024 Jul 22;52(13):7627-7649. doi: 10.1093/nar/gkae449. Nucleic Acids Res. 2024. PMID: 38813825 Free PMC article.
References
-
- Andrews S. 2010. FastQC: a Quality Control Tool for High Throughput Sequence Data, pp. Babraham Bioinformatics, Babraham Institute. United Kingdom: Cambridge.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
