New plasma diagnostic markers for colorectal cancer: transporter fragments of glutamate tRNA origin
- PMID: 38356701
- PMCID: PMC10861818
- DOI: 10.7150/jca.92102
New plasma diagnostic markers for colorectal cancer: transporter fragments of glutamate tRNA origin
Abstract
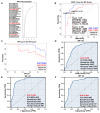
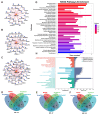
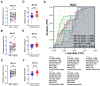
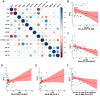
Colorectal cancer (CRC) is the second leading cause of cancer-related deaths worldwide. Early diagnosis of the disease can greatly improve the clinical prognosis for patients with CRC. Unfortunately, there are no current simple and effective early diagnostic markers available. The transfer RNA (tRNA)-derived RNA fragments (tRFs) are a class of small non-coding RNAs (sncRNAs), which have been shown to play an important role in the development and prognosis of CRC. However, only a few studies on tRFs as early diagnostic markers in CRC have been conducted. In this study, previously ignored tRFs expression data were extracted from six paired small RNA sequencing data in the Sequence Read Archive (SRA) database using MINTmap. Three i-tRFs, derived from the tRNA that transports glutamate (i-tRF-Glu), were identified and used to construct a random forest diagnostic model. The model performance was evaluated using the receiver operating characteristic (ROC) curve and precision-recall (PR) curve. The area under the curves (AUC) for the ROC and PR was 0.941 and 0.944, respectively. We further verified the differences in expression of the these i-tRF-Glu in the tissue and plasma of both CRC patients and healthy subjects using quantitative real-time PCR (qRT-PCR). We found that the ROC-AUC of the three was greater than traditional plasma tumor markers such as CEA and CA199. Our bioinformatics analysis suggested that the these i-tRF-Glu are associated with cancer development and glutamate (Glu)-glutamine (Gln) metabolism. Overall, our study uncovered these i-tRF-Glu that have early diagnostic significance and therapeutic potential for CRC, this warrants further investigation into the diagnostic and therapeutic potential of these i-tRF-Glu in CRC.
Keywords: colorectal cancer; glutamate metabolism; i-tRF; liquid biopsy; tRNA-derived fragments.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

Similar articles
-
Identifying Differentially Expressed tRNA-Derived Small Fragments as a Biomarker for the Progression and Metastasis of Colorectal Cancer.Dis Markers. 2022 Jan 6;2022:2646173. doi: 10.1155/2022/2646173. eCollection 2022. Dis Markers. 2022. PMID: 35035608 Free PMC article.
-
A novel tRNA-derived fragment tRF-3022b modulates cell apoptosis and M2 macrophage polarization via binding to cytokines in colorectal cancer.J Hematol Oncol. 2022 Dec 16;15(1):176. doi: 10.1186/s13045-022-01388-z. J Hematol Oncol. 2022. PMID: 36527118 Free PMC article.
-
5'-tRF-GlyGCC: a tRNA-derived small RNA as a novel biomarker for colorectal cancer diagnosis.Genome Med. 2021 Feb 9;13(1):20. doi: 10.1186/s13073-021-00833-x. Genome Med. 2021. PMID: 33563322 Free PMC article.
-
Extracellular vesicles-associated tRNA-derived fragments (tRFs): biogenesis, biological functions, and their role as potential biomarkers in human diseases.J Mol Med (Berl). 2022 May;100(5):679-695. doi: 10.1007/s00109-022-02189-0. Epub 2022 Mar 24. J Mol Med (Berl). 2022. PMID: 35322869 Free PMC article. Review.
-
The role of Transfer RNA-Derived Small RNAs (tsRNAs) in Digestive System Tumors.J Cancer. 2020 Oct 18;11(24):7237-7245. doi: 10.7150/jca.46055. eCollection 2020. J Cancer. 2020. PMID: 33193887 Free PMC article. Review.
Cited by
-
Computational Analysis Suggests That AsnGTT 3'-tRNA-Derived Fragments Are Potential Biomarkers in Papillary Thyroid Carcinoma.Int J Mol Sci. 2024 Oct 2;25(19):10631. doi: 10.3390/ijms251910631. Int J Mol Sci. 2024. PMID: 39408960 Free PMC article.
References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7–33. - PubMed
-
- Sung H, Ferlay J, Siegel RL. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Siegel RL, Miller KD, Goding Sauer A. et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–64. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

