Skp1 proteins are structural components of the synaptonemal complex in C. elegans
- PMID: 38354250
- PMCID: PMC10866564
- DOI: 10.1126/sciadv.adl4876
Skp1 proteins are structural components of the synaptonemal complex in C. elegans
Abstract
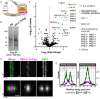
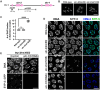
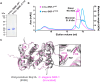
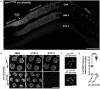
The synaptonemal complex (SC) is a zipper-like protein assembly that links homologous chromosomes to regulate recombination and segregation during meiosis. The SC has been notoriously refractory to in vitro reconstitution, thus leaving its molecular organization largely unknown. Here, we report a moonlighting function of two paralogous S-phase kinase-associated protein 1 (Skp1)-related proteins (SKR-1 and SKR-2), well-known adaptors of the Skp1-Cul1-F-box (SCF) ubiquitin ligase, as the key missing components of the SC in Caenorhabditis elegans. SKR proteins repurpose their SCF-forming interfaces to dimerize and interact with meiosis-specific SC proteins, thereby driving synapsis independent of SCF activity. SKR-1 enables the formation of the long-sought-after soluble complex with previously identified SC proteins in vitro, which we propose it to represent a complete SC building block. Our findings demonstrate how a conserved cell cycle regulator has been co-opted to interact with rapidly evolving meiotic proteins to construct the SC and provide a foundation for understanding its structure and assembly mechanisms.
Figures

Similar articles
-
Skp1 is a conserved structural component of the meiotic synaptonemal complex.bioRxiv [Preprint]. 2024 Jun 28:2024.06.24.600447. doi: 10.1101/2024.06.24.600447. bioRxiv. 2024. PMID: 38979327 Free PMC article. Preprint.
-
The structural role of Skp1 in the synaptonemal complex is conserved in nematodes.Genetics. 2024 Nov 6;228(3):iyae153. doi: 10.1093/genetics/iyae153. Genetics. 2024. PMID: 39293001 Free PMC article.
-
Multiple Skp1-related proteins in Caenorhabditis elegans: diverse patterns of interaction with Cullins and F-box proteins.Curr Biol. 2002 Feb 19;12(4):267-75. doi: 10.1016/s0960-9822(02)00657-7. Curr Biol. 2002. PMID: 11864566
-
Building the synaptonemal complex: Molecular interactions between the axis and the central region.PLoS Genet. 2023 Jul 20;19(7):e1010822. doi: 10.1371/journal.pgen.1010822. eCollection 2023 Jul. PLoS Genet. 2023. PMID: 37471284 Free PMC article. Review.
-
The synaptonemal complex of basal metazoan hydra: more similarities to vertebrate than invertebrate meiosis model organisms.J Genet Genomics. 2014 Mar 20;41(3):107-15. doi: 10.1016/j.jgg.2014.01.009. Epub 2014 Feb 20. J Genet Genomics. 2014. PMID: 24656231 Review.
Cited by
-
A suppressor screen in C. elegans identifies a multiprotein interaction that stabilizes the synaptonemal complex.Proc Natl Acad Sci U S A. 2023 Dec 12;120(50):e2314335120. doi: 10.1073/pnas.2314335120. Epub 2023 Dec 6. Proc Natl Acad Sci U S A. 2023. PMID: 38055743 Free PMC article.
-
Skp1 is a conserved structural component of the meiotic synaptonemal complex.bioRxiv [Preprint]. 2024 Jun 28:2024.06.24.600447. doi: 10.1101/2024.06.24.600447. bioRxiv. 2024. PMID: 38979327 Free PMC article. Preprint.
-
Establishment of the auxin inducible degron system for Babesia duncani: a conditional knockdown tool to study precise protein regulation in Babesia spp.Parasit Vectors. 2024 Oct 31;17(1):446. doi: 10.1186/s13071-024-06458-4. Parasit Vectors. 2024. PMID: 39478572 Free PMC article.
-
A suppressor screen in C. elegans identifies a multi-protein interaction interface that stabilizes the synaptonemal complex.bioRxiv [Preprint]. 2023 Aug 22:2023.08.21.554166. doi: 10.1101/2023.08.21.554166. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2023 Dec 12;120(50):e2314335120. doi: 10.1073/pnas.2314335120 PMID: 37662357 Free PMC article. Updated. Preprint.
-
The structural role of Skp1 in the synaptonemal complex is conserved in nematodes.Genetics. 2024 Nov 6;228(3):iyae153. doi: 10.1093/genetics/iyae153. Genetics. 2024. PMID: 39293001 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

