This is a preprint.
A large-scale cancer-specific protein-DNA interaction network
- PMID: 38352498
- PMCID: PMC10862707
- DOI: 10.1101/2024.01.24.577099
A large-scale cancer-specific protein-DNA interaction network
Update in
-
A large-scale cancer-specific protein-DNA interaction network.Life Sci Alliance. 2024 Jul 16;7(10):e202402641. doi: 10.26508/lsa.202402641. Print 2024 Oct. Life Sci Alliance. 2024. PMID: 39013578 Free PMC article.
Abstract
Cancer development and progression are generally associated with dysregulation of gene expression, often resulting from changes in transcription factor (TF) sequence or expression. Identifying key TFs involved in cancer gene regulation provides a framework for potential new therapeutics. This study presents a large-scale cancer gene TF-DNA interaction network as well as an extensive promoter clone resource for future studies. Most highly connected TFs do not show a preference for binding to promoters of genes associated with either good or poor cancer prognosis, suggesting that emerging strategies aimed at shifting gene expression balance between these two prognostic groups may be inherently complex. However, we identified potential for oncogene targeted therapeutics, with half of the tested oncogenes being potentially repressed by influencing specific activator or bifunctional TFs. Finally, we investigate the role of intrinsically disordered regions within the key cancer-related TF estrogen receptor ɑ (ESR1) on DNA binding and transcriptional activity, and found that these regions can have complex trade-offs in TF function. Altogether, our study not only broadens our knowledge of TFs involved in the cancer gene regulatory network but also provides a valuable resource for future studies, laying a foundation for potential therapeutic strategies targeting TFs in cancer.
Conflict of interest statement
Conflict of Interest The authors declare no conflicts of interest.
Figures

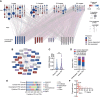
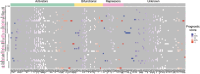

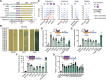
Similar articles
-
Atlas of regulated target genes of transcription factors (ART-TF) in human ES cells.BMC Bioinformatics. 2022 Sep 16;23(1):377. doi: 10.1186/s12859-022-04924-3. BMC Bioinformatics. 2022. PMID: 36114445 Free PMC article.
-
TF-Cluster: a pipeline for identifying functionally coordinated transcription factors via network decomposition of the shared coexpression connectivity matrix (SCCM).BMC Syst Biol. 2011 Apr 15;5:53. doi: 10.1186/1752-0509-5-53. BMC Syst Biol. 2011. PMID: 21496241 Free PMC article.
-
TF-centered downstream gene set enrichment analysis: Inference of causal regulators by integrating TF-DNA interactions and protein post-translational modifications information.BMC Bioinformatics. 2010 Dec 14;11 Suppl 11(Suppl 11):S5. doi: 10.1186/1471-2105-11-S11-S5. BMC Bioinformatics. 2010. PMID: 21172055 Free PMC article.
-
Transcription factors in the development and treatment of immune disorders.Transcription. 2023 Dec 15:1-23. doi: 10.1080/21541264.2023.2294623. Online ahead of print. Transcription. 2023. PMID: 38100543 Review.
-
Chicken ovalbumin upstream promoter-transcription factors and their regulation.J Steroid Biochem Mol Biol. 1996 Jan;56(1-6 Spec No):81-5. doi: 10.1016/0960-0760(95)00225-1. J Steroid Biochem Mol Biol. 1996. PMID: 8603050 Review.
References
-
- Djakiew D. (2000) Dysregulated expression of growth factors and their receptors in the development of prostate cancer. Prostate, 42, 150–160. - PubMed
-
- Vervoort S.J., Devlin J.R., Kwiatkowski N., Teng M., Gray N.S. and Johnstone R.W. (2022) Targeting transcription cycles in cancer. Nat Rev Cancer, 22, 5–24. - PubMed
-
- Hanahan D. and Weinberg R.A. (2011) Hallmarks of cancer: the next generation. Cell, 144, 646–674. - PubMed
-
- Sjostedt E., Zhong W., Fagerberg L., Karlsson M., Mitsios N., Adori C., Oksvold P., Edfors F., Limiszewska A., Hikmet F. et al. (2020) An atlas of the protein-coding genes in the human, pig, and mouse brain. Science, 367. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
