The molecular machinery of meiotic recombination
- PMID: 38348856
- PMCID: PMC10903461
- DOI: 10.1042/BST20230712
The molecular machinery of meiotic recombination
Abstract
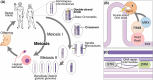
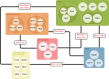
Meiotic recombination, a cornerstone of eukaryotic diversity and individual genetic identity, is essential for the creation of physical linkages between homologous chromosomes, facilitating their faithful segregation during meiosis I. This process requires that germ cells generate controlled DNA lesions within their own genome that are subsequently repaired in a specialised manner. Repair of these DNA breaks involves the modulation of existing homologous recombination repair pathways to generate crossovers between homologous chromosomes. Decades of genetic and cytological studies have identified a multitude of factors that are involved in meiotic recombination. Recent work has started to provide additional mechanistic insights into how these factors interact with one another, with DNA, and provide the molecular outcomes required for a successful meiosis. Here, we provide a review of the recent developments with a focus on protein structures and protein-protein interactions.
Keywords: DNA replication and recombination; chromosomes; meiosis.
© 2024 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

Similar articles
-
The meiotic-specific Mek1 kinase in budding yeast regulates interhomolog recombination and coordinates meiotic progression with double-strand break repair.Curr Genet. 2019 Jun;65(3):631-641. doi: 10.1007/s00294-019-00937-3. Epub 2019 Jan 22. Curr Genet. 2019. PMID: 30671596 Free PMC article. Review.
-
Alignment of Homologous Chromosomes and Effective Repair of Programmed DNA Double-Strand Breaks during Mouse Meiosis Require the Minichromosome Maintenance Domain Containing 2 (MCMDC2) Protein.PLoS Genet. 2016 Oct 19;12(10):e1006393. doi: 10.1371/journal.pgen.1006393. eCollection 2016 Oct. PLoS Genet. 2016. PMID: 27760146 Free PMC article.
-
Structural maintenance of chromosomes (SMC) proteins promote homolog-independent recombination repair in meiosis crucial for germ cell genomic stability.PLoS Genet. 2010 Jul 22;6(7):e1001028. doi: 10.1371/journal.pgen.1001028. PLoS Genet. 2010. PMID: 20661436 Free PMC article.
-
Kinetochores, cohesin, and DNA breaks: Controlling meiotic recombination within pericentromeres.Yeast. 2019 Mar;36(3):121-127. doi: 10.1002/yea.3366. Epub 2019 Feb 3. Yeast. 2019. PMID: 30625250 Free PMC article. Review.
-
Gene conversion: a non-Mendelian process integral to meiotic recombination.Heredity (Edinb). 2022 Jul;129(1):56-63. doi: 10.1038/s41437-022-00523-3. Epub 2022 Apr 7. Heredity (Edinb). 2022. PMID: 35393552 Free PMC article.
Cited by
-
Proximity labeling reveals new functional relationships between meiotic recombination proteins in S. cerevisiae.PLoS Genet. 2024 Oct 15;20(10):e1011432. doi: 10.1371/journal.pgen.1011432. eCollection 2024 Oct. PLoS Genet. 2024. PMID: 39405359 Free PMC article.
-
The chromatin-associated 53BP1 ortholog, HSR-9, regulates recombinational repair and X chromosome segregation in the Caenorhabditis elegans germ line.bioRxiv [Preprint]. 2024 Apr 15:2024.04.12.589267. doi: 10.1101/2024.04.12.589267. bioRxiv. 2024. Update in: Genetics. 2024 Aug 7;227(4):iyae102. doi: 10.1093/genetics/iyae102 PMID: 38659880 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

