An angiogenesis-associated gene-based signature predicting prognosis and immunotherapy efficacy of head and neck squamous cell carcinoma patients
- PMID: 38347320
- PMCID: PMC10861726
- DOI: 10.1007/s00432-024-05606-8
An angiogenesis-associated gene-based signature predicting prognosis and immunotherapy efficacy of head and neck squamous cell carcinoma patients
Abstract
Objectives: To develop a model that can assist in the diagnosis and prediction of prognosis for head and neck squamous cell carcinoma (HNSCC).
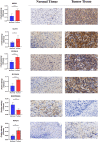
Materials and methods: Data from TCGA and GEO databases were used to generate normalized gene expression data. Consensus Cluster Plus was used for cluster analysis and the relationship between angiogenesis-associated gene (AAG) expression patterns, clinical characteristics and survival was examined. Support vector machine (SVM) and least absolute shrinkage and selection operator (LASSO) analyzes and multiple logistic regression analyzes were performed to determine the diagnostic model, and a prognostic nomogram was constructed using univariate and multivariate Cox regression analyses. ESTIMATE, XCELL, TIMER, QUANTISEQ, MCPCOUNTER, EPIC, CIBERSORT-ABS, CIBERSORT algorithms were used to assess the immune microenvironment of HNSCC patients. In addition, gene set enrichment analysis, treatment sensitivity analysis, and AAGs mutation studies were performed. Finally, we also performed immunohistochemistry (IHC) staining in the tissue samples.
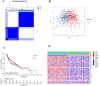
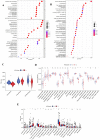
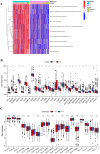
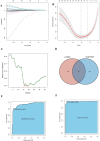
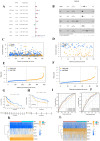
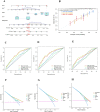
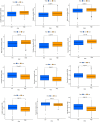
Results: We classified HNSCC patients into subtypes based on differences in AAG expression from TCGA and GEO databases. There are differences in clinical features, TME, and immune-related gene expression between two subgroups. We constructed a HNSCC diagnostic model based on nine AAGs, which has good sensitivity and specificity. After further screening, we constructed a prognostic risk signature for HNSCC based on six AAGs. The constructed risk score had a good independent prognostic significance, and it was further constructed into a prognostic nomogram together with age and stage. Different prognostic risk groups have differences in immune microenvironment, drug sensitivity, gene enrichment and gene mutation.
Conclusion: We have constructed a diagnostic and prognostic model for HNSCC based on AAG, which has good performance. The constructed prognostic risk score is closely related to tumor immune microenvironment and immunotherapy response.
Keywords: Angiogenesis-associated gene; Diagnosis; HNSCC; Immunotherapy; Prognostic signature.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
A meta-validated immune infiltration-related gene model predicts prognosis and immunotherapy sensitivity in HNSCC.BMC Cancer. 2023 Jan 13;23(1):45. doi: 10.1186/s12885-023-10532-y. BMC Cancer. 2023. PMID: 36639648 Free PMC article.
-
Development of Chromatin Regulator-related Molecular Subtypes and a Signature to Predict Prognosis and Immunotherapeutic Response in Head and Neck Squamous Cell Carcinoma.Curr Cancer Drug Targets. 2024;24(8):804-819. doi: 10.2174/0115680096274798231121053634. Curr Cancer Drug Targets. 2024. PMID: 38310463 Free PMC article.
-
A novel Pyroptosis-related long non-coding RNA signature for predicting the prognosis and immune landscape of head and neck squamous cell carcinoma.Cancer Med. 2022 Dec;11(24):5097-5112. doi: 10.1002/cam4.4819. Epub 2022 May 14. Cancer Med. 2022. PMID: 35567376 Free PMC article. Clinical Trial.
-
Development and experimental verification of a prognosis model for disulfidptosis-associated genes in HNSCC.Medicine (Baltimore). 2024 Mar 22;103(12):e37308. doi: 10.1097/MD.0000000000037308. Medicine (Baltimore). 2024. PMID: 38518012 Free PMC article. Review.
-
Mouse Models for Head and Neck Squamous Cell Carcinoma.J Dent Res. 2024 Jun;103(6):585-595. doi: 10.1177/00220345241240997. Epub 2024 May 9. J Dent Res. 2024. PMID: 38722077 Review.
Cited by
-
An anoikis-related lncRNA signature may predict the prognosis, immune infiltration, and drug sensitivity in esophageal cancer.Heliyon. 2024 May 14;10(10):e31202. doi: 10.1016/j.heliyon.2024.e31202. eCollection 2024 May 30. Heliyon. 2024. PMID: 38803953 Free PMC article.
References
-
- Bejarano L, Jordāo MJC, Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Discov. 2021;11:933–959. doi: 10.1158/2159-8290.Cd-20-1808. - DOI - PubMed
-
- Butkiewicz D, Gdowicz-Klosok A, Krzesniak M, Rutkowski T, Krzywon A, Cortez AJ, Dominczyk I, Skladowski K. Association of genetic variants in ANGPT/TEK and VEGF/VEGFR with progression and survival in head and neck squamous cell carcinoma treated with radiotherapy or radiochemotherapy. Cancers (basel) 2020 doi: 10.3390/cancers12061506. - DOI - PMC - PubMed
-
- Chalabi M, Fanchi LF, Dijkstra KK, Van den Berg JG, Aalbers AG, Sikorska K, Lopez-Yurda M, Grootscholten C, Beets GL, Snaebjornsson P, et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat Med. 2020;26:566–576. doi: 10.1038/s41591-020-0805-8. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

