PREDAC-CNN: predicting antigenic clusters of seasonal influenza A viruses with convolutional neural network
- PMID: 38343322
- PMCID: PMC10859661
- DOI: 10.1093/bib/bbae033
PREDAC-CNN: predicting antigenic clusters of seasonal influenza A viruses with convolutional neural network
Abstract
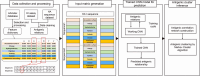
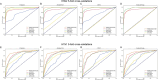
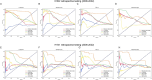
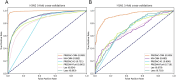
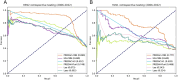
Vaccination stands as the most effective and economical strategy for prevention and control of influenza. The primary target of neutralizing antibodies is the surface antigen hemagglutinin (HA). However, ongoing mutations in the HA sequence result in antigenic drift. The success of a vaccine is contingent on its antigenic congruence with circulating strains. Thus, predicting antigenic variants and deducing antigenic clusters of influenza viruses are pivotal for recommendation of vaccine strains. The antigenicity of influenza A viruses is determined by the interplay of amino acids in the HA1 sequence. In this study, we exploit the ability of convolutional neural networks (CNNs) to extract spatial feature representations in the convolutional layers, which can discern interactions between amino acid sites. We introduce PREDAC-CNN, a model designed to track antigenic evolution of seasonal influenza A viruses. Accessible at http://predac-cnn.cloudna.cn, PREDAC-CNN formulates a spatially oriented representation of the HA1 sequence, optimized for the convolutional framework. It effectively probes interactions among amino acid sites in the HA1 sequence. Also, PREDAC-CNN focuses exclusively on physicochemical attributes crucial for the antigenicity of influenza viruses, thereby eliminating unnecessary amino acid embeddings. Together, PREDAC-CNN is adept at capturing interactions of amino acid sites within the HA1 sequence and examining the collective impact of point mutations on antigenic variation. Through 5-fold cross-validation and retrospective testing, PREDAC-CNN has shown superior performance in predicting antigenic variants compared to its counterparts. Additionally, PREDAC-CNN has been instrumental in identifying predominant antigenic clusters for A/H3N2 (1968-2023) and A/H1N1 (1977-2023) viruses, significantly aiding in vaccine strain recommendation.
Keywords: HA sequence; antigenic cluster; antigenic variant; convolutional neural network; seasonal influenza A viruses.
© The Author(s) 2024. Published by Oxford University Press.
Figures


Similar articles
-
Mapping of H3N2 influenza antigenic evolution in China reveals a strategy for vaccine strain recommendation.Nat Commun. 2012 Feb 28;3:709. doi: 10.1038/ncomms1710. Nat Commun. 2012. PMID: 22426230
-
PREDAC-H3: a user-friendly platform for antigenic surveillance of human influenza a(H3N2) virus based on hemagglutinin sequences.Bioinformatics. 2016 Aug 15;32(16):2526-7. doi: 10.1093/bioinformatics/btw185. Epub 2016 Apr 12. Bioinformatics. 2016. PMID: 27153622
-
Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. swine.J Virol. 2014 May;88(9):4752-63. doi: 10.1128/JVI.03805-13. Epub 2014 Feb 12. J Virol. 2014. PMID: 24522915 Free PMC article.
-
Genetic characterization of influenza viruses from influenza-related hospital admissions in the St. Petersburg and Valencia sites of the Global Influenza Hospital Surveillance Network during the 2013/14 influenza season.J Clin Virol. 2016 Nov;84:32-38. doi: 10.1016/j.jcv.2016.09.006. Epub 2016 Sep 28. J Clin Virol. 2016. PMID: 27690141
-
Genetic diversity of influenza A viruses circulating in Bulgaria during the 2018-2019 winter season.J Med Microbiol. 2020 Jul;69(7):986-998. doi: 10.1099/jmm.0.001198. J Med Microbiol. 2020. PMID: 32459617 Free PMC article.
Cited by
-
MetaFluAD: meta-learning for predicting antigenic distances among influenza viruses.Brief Bioinform. 2024 Jul 25;25(5):bbae395. doi: 10.1093/bib/bbae395. Brief Bioinform. 2024. PMID: 39129362 Free PMC article.
References
-
- Influenza Fact Sheet WHO. https://www.who.int/en/news-room/fact-sheets/detail/influenza-(seasonal).
-
- Virelizier J-L. Host defenses against influenza virus: the role of anti-hemagglutinin antibody. J Immunol 1975;115:434–9. - PubMed
-
- Cox NJ, Lynnette Brammer T, Regnery HL. Influenza: global surveillance for epidemic and pandemic variants. Eur J Epidemiol 1994;10:467–70. - PubMed
MeSH terms
Substances
Grants and funding
- 32070678/National Natural Science Foundation of China
- BK20220278/Natural Science Foundation of Jiangsu Province
- EKPG21-12/Emergency Key Program of Guangzhou Laboratory
- 2021YFC2302000/National Key Research and Development Program of China
- 2022-1G-1131/Capital's Funds for Health Improvement and Research
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

