Mechanism of DNA Intercalation by Chloroquine Provides Insights into Toxicity
- PMID: 38338688
- PMCID: PMC10855526
- DOI: 10.3390/ijms25031410
Mechanism of DNA Intercalation by Chloroquine Provides Insights into Toxicity
Abstract
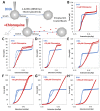
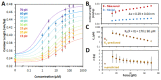
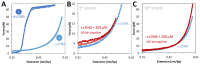
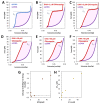
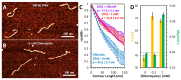
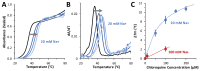
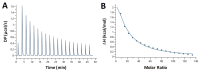
Chloroquine has been used as a potent antimalarial, anticancer drug, and prophylactic. While chloroquine is known to interact with DNA, the details of DNA-ligand interactions have remained unclear. Here we characterize chloroquine-double-stranded DNA binding with four complementary approaches, including optical tweezers, atomic force microscopy, duplex DNA melting measurements, and isothermal titration calorimetry. We show that chloroquine intercalates into double stranded DNA (dsDNA) with a KD ~ 200 µM, and this binding is entropically driven. We propose that chloroquine-induced dsDNA intercalation, which happens in the same concentration range as its observed toxic effects on cells, is responsible for the drug's cytotoxicity.
Keywords: AFM; DNA binding; DNA melting; chloroquine; intercalation; isothermal titration calorimetry; optical tweezers; single molecule.
Conflict of interest statement
The authors declare no conflicts of interest, and the funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
Similar articles
-
Amodiaquine Nonspecifically Binds Double Stranded and Three-Way Junction DNA Structures.Chembiochem. 2024 Jul 15;25(14):e202400116. doi: 10.1002/cbic.202400116. Epub 2024 Jun 16. Chembiochem. 2024. PMID: 38668388
-
Interaction of an anticancer benzopyrane derivative with DNA: Biophysical, biochemical, and molecular modeling studies.Biochim Biophys Acta Gen Subj. 2023 Jun;1867(6):130347. doi: 10.1016/j.bbagen.2023.130347. Epub 2023 Mar 21. Biochim Biophys Acta Gen Subj. 2023. PMID: 36958685
-
Interaction of chloroquine and its analogues with heme: An isothermal titration calorimetric study.Biochem Biophys Res Commun. 2000 Oct 5;276(3):1075-9. doi: 10.1006/bbrc.2000.3592. Biochem Biophys Res Commun. 2000. PMID: 11027592
-
Biophysical characterization of DNA binding from single molecule force measurements.Phys Life Rev. 2010 Sep;7(3):299-341. doi: 10.1016/j.plrev.2010.06.001. Epub 2010 Jun 4. Phys Life Rev. 2010. PMID: 20576476 Free PMC article. Review.
-
Chloroquine-based hybrid molecules as promising novel chemotherapeutic agents.Eur J Pharmacol. 2015 Sep 5;762:472-86. doi: 10.1016/j.ejphar.2015.04.048. Epub 2015 May 8. Eur J Pharmacol. 2015. PMID: 25959387 Review.
References
-
- U.S. Food & Drug Administration Coronavirus (COVID-19) Update: FDA Revokes Emergency Use Authorization for Chloroquine and Hydroxychloroquine [Press Release] [(accessed on 28 December 2023)]; Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19....
-
- Yang J., Guo Z., Liu X., Liu Q., Wu M., Yao X., Liu Y., Cui C., Li H., Song C., et al. Cytotoxicity Evaluation of Chloroquine and Hydroxychloroquine in Multiple Cell Lines and Tissues by Dynamic Imaging System and Physiologically Based Pharmacokinetic Model. Front. Pharmacol. 2020;11:574720. doi: 10.3389/fphar.2020.574720. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

