Snowprint: a predictive tool for genetic biosensor discovery
- PMID: 38336860
- PMCID: PMC10858194
- DOI: 10.1038/s42003-024-05849-8
Snowprint: a predictive tool for genetic biosensor discovery
Abstract
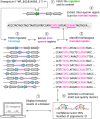
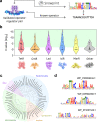
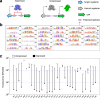
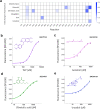
Bioengineers increasingly rely on ligand-inducible transcription regulators for chemical-responsive control of gene expression, yet the number of regulators available is limited. Novel regulators can be mined from genomes, but an inadequate understanding of their DNA specificity complicates genetic design. Here we present Snowprint, a simple yet powerful bioinformatic tool for predicting regulator:operator interactions. Benchmarking results demonstrate that Snowprint predictions are significantly similar for >45% of experimentally validated regulator:operator pairs from organisms across nine phyla and for regulators that span five distinct structural families. We then use Snowprint to design promoters for 33 previously uncharacterized regulators sourced from diverse phylogenies, of which 28 are shown to influence gene expression and 24 produce a >20-fold dynamic range. A panel of the newly repurposed regulators are then screened for response to biomanufacturing-relevant compounds, yielding new sensors for a polyketide (olivetolic acid), terpene (geraniol), steroid (ursodiol), and alkaloid (tetrahydropapaverine) with induction ratios up to 10.7-fold. Snowprint represents a unique, protein-agnostic tool that greatly facilitates the discovery of ligand-inducible transcriptional regulators for bioengineering applications. A web-accessible version of Snowprint is available at https://snowprint.groov.bio .
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Far Posterior Approach for Rib Fracture Fixation: Surgical Technique and Tips.JBJS Essent Surg Tech. 2024 Dec 6;14(4):e23.00094. doi: 10.2106/JBJS.ST.23.00094. eCollection 2024 Oct-Dec. JBJS Essent Surg Tech. 2024. PMID: 39650795 Free PMC article.
-
Enabling Systemic Identification and Functionality Profiling for Cdc42 Homeostatic Modulators.bioRxiv [Preprint]. 2024 Jan 8:2024.01.05.574351. doi: 10.1101/2024.01.05.574351. bioRxiv. 2024. Update in: Commun Chem. 2024 Nov 19;7(1):271. doi: 10.1038/s42004-024-01352-7. PMID: 38260445 Free PMC article. Updated. Preprint.
-
Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article.
-
Mobile apps to reduce depressive symptoms and alcohol use in youth: A systematic review and meta-analysis: A systematic review.Campbell Syst Rev. 2024 Apr 26;20(2):e1398. doi: 10.1002/cl2.1398. eCollection 2024 Jun. Campbell Syst Rev. 2024. PMID: 38680950 Free PMC article. Review.
Cited by
-
Synthetic genomes unveil the effects of synonymous recoding.bioRxiv [Preprint]. 2024 Jun 16:2024.06.16.599206. doi: 10.1101/2024.06.16.599206. bioRxiv. 2024. PMID: 38915524 Free PMC article. Preprint.
-
Ligify: Automated Genome Mining for Ligand-Inducible Transcription Factors.ACS Synth Biol. 2024 Aug 16;13(8):2577-2586. doi: 10.1021/acssynbio.4c00372. Epub 2024 Jul 19. ACS Synth Biol. 2024. PMID: 39029917
-
Microbial biosensors for diagnostics, surveillance and epidemiology: Today's achievements and tomorrow's prospects.Microb Biotechnol. 2024 Nov;17(11):e70047. doi: 10.1111/1751-7915.70047. Microb Biotechnol. 2024. PMID: 39548716 Free PMC article. Review.
-
Accelerating Genetic Sensor Development, Scale-up, and Deployment Using Synthetic Biology.Biodes Res. 2024 Jun 25;6:0037. doi: 10.34133/bdr.0037. eCollection 2024. Biodes Res. 2024. PMID: 38919711 Free PMC article.
-
In vitro transcription-based biosensing of glycolate for prototyping of a complex enzyme cascade.Synth Biol (Oxf). 2024 Sep 20;9(1):ysae013. doi: 10.1093/synbio/ysae013. eCollection 2024. Synth Biol (Oxf). 2024. PMID: 39399720 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

