Distinct cellular expression and subcellular localization of Kv2 voltage-gated K+ channel subtypes in dorsal root ganglion neurons conserved between mice and humans
- PMID: 38335058
- PMCID: PMC10861167
- DOI: 10.1002/cne.25575
Distinct cellular expression and subcellular localization of Kv2 voltage-gated K+ channel subtypes in dorsal root ganglion neurons conserved between mice and humans
Abstract
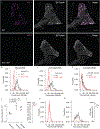
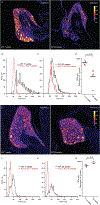
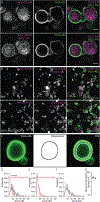
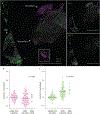
The distinct organization of Kv2 voltage-gated potassium channels on and near the cell body of brain neurons enables their regulation of action potentials and specialized membrane contact sites. Somatosensory neurons have a pseudounipolar morphology and transmit action potentials from peripheral nerve endings through axons that bifurcate to the spinal cord and the cell body within ganglia including the dorsal root ganglia (DRG). Kv2 channels regulate action potentials in somatosensory neurons, yet little is known about where Kv2 channels are located. Here, we define the cellular and subcellular localization of the Kv2 paralogs, Kv2.1 and Kv2.2, in DRG somatosensory neurons with a panel of antibodies, cell markers, and genetically modified mice. We find that relative to spinal cord neurons, DRG neurons have similar levels of detectable Kv2.1 and higher levels of Kv2.2. In older mice, detectable Kv2.2 remains similar, while detectable Kv2.1 decreases. Both Kv2 subtypes adopt clustered subcellular patterns that are distinct from central neurons. Most DRG neurons co-express Kv2.1 and Kv2.2, although neuron subpopulations show preferential expression of Kv2.1 or Kv2.2. We find that Kv2 protein expression and subcellular localization are similar between mouse and human DRG neurons. We conclude that the organization of both Kv2 channels is consistent with physiological roles in the somata and stem axons of DRG neurons. The general prevalence of Kv2.2 in DRG as compared to central neurons and the enrichment of Kv2.2 relative to detectable Kv2.1 in older mice, proprioceptors, and axons suggest more widespread roles for Kv2.2 in DRG neurons.
Keywords: Kv2.1; Kv2.2; dorsal root ganglion; somatosensory neurons; voltage-gated ion channels.
© 2024 Wiley Periodicals LLC.
Conflict of interest statement
Figures


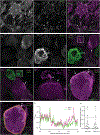

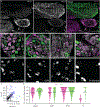
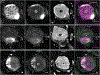
Update of
-
Distinct cellular expression and subcellular localization of Kv2 voltage-gated K+ channel subtypes in dorsal root ganglion neurons conserved between mice and humans.bioRxiv [Preprint]. 2023 Dec 24:2023.03.01.530679. doi: 10.1101/2023.03.01.530679. bioRxiv. 2023. Update in: J Comp Neurol. 2024 Feb;532(2):e25575. doi: 10.1002/cne.25575. PMID: 38187582 Free PMC article. Updated. Preprint.
Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Ih current stabilizes excitability in rodent DRG neurons and reverses hyperexcitability in a nociceptive neuron model of inherited neuropathic pain.J Physiol. 2023 Dec;601(23):5341-5366. doi: 10.1113/JP284999. Epub 2023 Oct 17. J Physiol. 2023. PMID: 37846879 Free PMC article.
-
Distinct cellular expression and subcellular localization of Kv2 voltage-gated K+ channel subtypes in dorsal root ganglion neurons conserved between mice and humans.bioRxiv [Preprint]. 2023 Dec 24:2023.03.01.530679. doi: 10.1101/2023.03.01.530679. bioRxiv. 2023. Update in: J Comp Neurol. 2024 Feb;532(2):e25575. doi: 10.1002/cne.25575. PMID: 38187582 Free PMC article. Updated. Preprint.
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
-
Kv2 channels do not function as canonical delayed rectifiers in spinal motoneurons.iScience. 2024 Jul 3;27(8):110444. doi: 10.1016/j.isci.2024.110444. eCollection 2024 Aug 16. iScience. 2024. PMID: 39148717 Free PMC article.
References
-
- Bishop HI, Guan D, Bocksteins E, Parajuli LK, Murray KD, Cobb MM, … Trimmer JS (2015). Distinct cell- and layer-specific expression patterns and independent regulation of Kv2 channel subtypes in cortical pyramidal neurons. J Neurosci, 35(44), 14922–14942. doi:10.1523/jneurosci.1897-15.2015 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

