Predicting horizontal gene transfers with perfect transfer networks
- PMID: 38321476
- PMCID: PMC10848447
- DOI: 10.1186/s13015-023-00242-2
Predicting horizontal gene transfers with perfect transfer networks
Abstract
Background: Horizontal gene transfer inference approaches are usually based on gene sequences: parametric methods search for patterns that deviate from a particular genomic signature, while phylogenetic methods use sequences to reconstruct the gene and species trees. However, it is well-known that sequences have difficulty identifying ancient transfers since mutations have enough time to erase all evidence of such events. In this work, we ask whether character-based methods can predict gene transfers. Their advantage over sequences is that homologous genes can have low DNA similarity, but still have retained enough important common motifs that allow them to have common character traits, for instance the same functional or expression profile. A phylogeny that has two separate clades that acquired the same character independently might indicate the presence of a transfer even in the absence of sequence similarity.
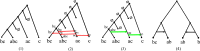
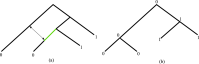
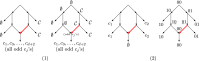
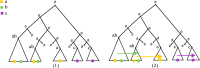
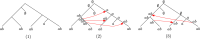
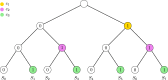
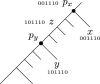
Our contributions: We introduce perfect transfer networks, which are phylogenetic networks that can explain the character diversity of a set of taxa under the assumption that characters have unique births, and that once a character is gained it is rarely lost. Examples of such traits include transposable elements, biochemical markers and emergence of organelles, just to name a few. We study the differences between our model and two similar models: perfect phylogenetic networks and ancestral recombination networks. Our goals are to initiate a study on the structural and algorithmic properties of perfect transfer networks. We then show that in polynomial time, one can decide whether a given network is a valid explanation for a set of taxa, and show how, for a given tree, one can add transfer edges to it so that it explains a set of taxa. We finally provide lower and upper bounds on the number of transfers required to explain a set of taxa, in the worst case.
Keywords: Character-based; Gene-expression; Horizontal gene transfer; Indirect phylogenetic methods; Perfect phylogenies; Tree-based networks.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Phylogenetic Trees and Networks Reduce to Phylogenies on Binary States: Does It Furnish an Explanation to the Robustness of Phylogenetic Trees against Lateral Transfers.Evol Bioinform Online. 2015 Oct 13;11:213-21. doi: 10.4137/EBO.S28158. eCollection 2015. Evol Bioinform Online. 2015. PMID: 26508826 Free PMC article.
-
A decomposition theory for phylogenetic networks and incompatible characters.J Comput Biol. 2007 Dec;14(10):1247-72. doi: 10.1089/cmb.2006.0137. J Comput Biol. 2007. PMID: 18047426 Free PMC article.
-
Invariant transformers of Robinson and Foulds distance matrices for Convolutional Neural Network.J Bioinform Comput Biol. 2022 Aug;20(4):2250012. doi: 10.1142/S0219720022500123. Epub 2022 Jul 6. J Bioinform Comput Biol. 2022. PMID: 35798684
-
The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification.Int J Syst Evol Microbiol. 2002 Jan;52(Pt 1):7-76. doi: 10.1099/00207713-52-1-7. Int J Syst Evol Microbiol. 2002. PMID: 11837318 Review.
-
Protein based molecular markers provide reliable means to understand prokaryotic phylogeny and support Darwinian mode of evolution.Front Cell Infect Microbiol. 2012 Jul 26;2:98. doi: 10.3389/fcimb.2012.00098. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919687 Free PMC article. Review.
References
-
- Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3(9):711–721. - PubMed
-
- Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9(8):605–618. - PubMed
-
- Irwin NA, Pittis AA, Richards TA, Keeling PJ. Systematic evaluation of horizontal gene transfer between eukaryotes and viruses. Nat Microbiol. 2022;7(2):327–336. - PubMed
LinkOut - more resources
Full Text Sources

