FSTL3 promotes tumor immune evasion and attenuates response to anti-PD1 therapy by stabilizing c-Myc in colorectal cancer
- PMID: 38302412
- PMCID: PMC10834545
- DOI: 10.1038/s41419-024-06469-0
FSTL3 promotes tumor immune evasion and attenuates response to anti-PD1 therapy by stabilizing c-Myc in colorectal cancer
Abstract
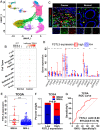
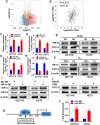
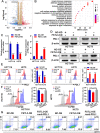
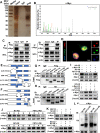
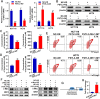
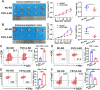
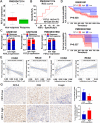
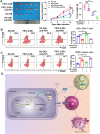
Programmed cell death 1 ligand 1 (PDL1)/programmed cell death 1 (PD1) blockade immunotherapy provides a prospective strategy for the treatment of colorectal cancer (CRC), but various constraints on the effectiveness of the treatment are still remaining. As reported in previous studies, follistatin-like 3 (FSTL3) could mediate inflammatory response in macrophages by induction lipid accumulation. Herein, we revealed that FSTL3 were overexpressed in malignant cells in the CRC microenvironment, notably, the expression level of FSTL3 was related to tumor immune evasion and the clinical efficacy of anti-PD1 therapy. Further studies determined that hypoxic tumor microenvironment induced the FSTL3 expression via HIF1α in CRC cells, FSTL3 could bind to the transcription factor c-Myc (354-406 amino acids) to suppress the latter's ubiquitination and increase its stability, thereby to up-regulated the expression of PDL1 and indoleamine 2,3-dioxygenase 1 (IDO1). The results in the immunocompetent tumor models verified that FSLT3 knockout in tumor cells increased the proportion of CD8+ T cells in the tumor microenvironment, reduced the proportion of regulatory T cells (CD25+ Foxp3+) and exhausted T cells (PD1+ CD8+), and synergistically improved the anti-PD1 therapy efficacy. To sum up, FSTL3 enhanced c-Myc-mediated transcriptional regulation to promote immune evasion and attenuates response to anti-PD1 therapy in CRC, suggesting the potential of FSTL3 as a biomarker of immunotherapeutic efficacy as well as a novel immunotherapeutic target in CRC.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Follistatin-Like 3 Correlates With Lymph Node Metastasis and Serves as a Biomarker of Extracellular Matrix Remodeling in Colorectal Cancer.Front Immunol. 2021 Jul 16;12:717505. doi: 10.3389/fimmu.2021.717505. eCollection 2021. Front Immunol. 2021. PMID: 34335633 Free PMC article.
-
Immunogenomics of Colorectal Cancer Response to Checkpoint Blockade: Analysis of the KEYNOTE 177 Trial and Validation Cohorts.Gastroenterology. 2021 Oct;161(4):1179-1193. doi: 10.1053/j.gastro.2021.06.064. Epub 2021 Jun 29. Gastroenterology. 2021. PMID: 34197832 Free PMC article.
-
Lactobacillus gallinarum-derived metabolites boost anti-PD1 efficacy in colorectal cancer by inhibiting regulatory T cells through modulating IDO1/Kyn/AHR axis.Gut. 2023 Nov 24;72(12):2272-2285. doi: 10.1136/gutjnl-2023-329543. Gut. 2023. PMID: 37770127 Free PMC article.
-
Involvement of tumor immune microenvironment metabolic reprogramming in colorectal cancer progression, immune escape, and response to immunotherapy.Front Immunol. 2024 Jul 25;15:1353787. doi: 10.3389/fimmu.2024.1353787. eCollection 2024. Front Immunol. 2024. PMID: 39119332 Free PMC article. Review.
-
PD-1/PD-L1 Checkpoint Inhibitors in Tumor Immunotherapy.Front Pharmacol. 2021 Sep 1;12:731798. doi: 10.3389/fphar.2021.731798. eCollection 2021. Front Pharmacol. 2021. PMID: 34539412 Free PMC article. Review.
Cited by
-
Effect of colorectal cancer stem cells on the development and metastasis of colorectal cancer.World J Gastrointest Oncol. 2024 Nov 15;16(11):4354-4368. doi: 10.4251/wjgo.v16.i11.4354. World J Gastrointest Oncol. 2024. PMID: 39554751 Free PMC article. Review.
-
Ubiquitin modification in the regulation of tumor immunotherapy resistance mechanisms and potential therapeutic targets.Exp Hematol Oncol. 2024 Aug 30;13(1):91. doi: 10.1186/s40164-024-00552-0. Exp Hematol Oncol. 2024. PMID: 39223632 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

