Repurposing proteasome inhibitors for improved treatment of triple-negative breast cancer
- PMID: 38286854
- PMCID: PMC10825133
- DOI: 10.1038/s41420-024-01819-5
Repurposing proteasome inhibitors for improved treatment of triple-negative breast cancer
Abstract
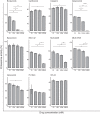
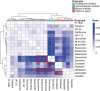
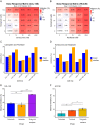
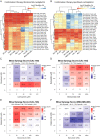
Triple-negative breast cancer (TNBC) is associated with poor prognosis and limited treatment options due to the lack of important receptors (estrogen receptor [ER], progesterone receptor [PR], and human epidermal growth factor receptor 2 [HER2]) used for targeted therapy. However, high-throughput in vitro drug screening of cell lines is a powerful tool for identifying effective drugs for a disease. Here, we determine the intrinsic chemosensitivity of TNBC cell lines to proteasome inhibitors (PIs), thereby identifying potentially potent 2-drug combinations for TNBC. Eight TNBC cell lines (BT-549, CAL-148, HCC1806, HCC38, HCC70, MDA-MB-436, MDA-MB-453, and MDA-MB-468) and two controls (MCF-10A and MCF-7) were first exposed to 18 drugs (11 PIs and 7 clinically relevant chemotherapeutic agents) as monotherapy, followed by prediction of potent 2-drug combinations using the IDACombo pipeline. The synergistic effects of the 2-drug combinations were evaluated with SynergyFinder in four TNBC cell lines (CAL-148, HCC1806, HCC38, and MDA-MB-468) and three controls (BT-474, MCF-7, and T47D) in vitro, followed by further evaluation of tumor regression in zebrafish tumor models established using HCC1806 and MCF-7 cells. Monotherapy identified nine effective drugs (bortezomib, carfilzomib, cisplatin, delanzomib, docetaxel, epoxomicin, MLN-2238, MLN-9708, and nedaplatin) across all cell lines. PIs (e.g., bortezomib, delanzomib, and epoxomicin) were highly potent drugs in TNBC cells, of which bortezomib and delanzomib inhibited the chymotrypsin-like activity of the 20 S proteasome by 100% at 10 µM. Moreover, several potent 2-drug combinations (e.g., bortezomib+nedaplatin and epoxomicin+epirubicin) that killed virtually 100% of cells were also identified. Although HCC1806- and MCF-7-derived xenografts treated with bortezomib+nedaplatin and carboplatin+paclitaxel were smaller, HCC1806 cells frequently metastasized to the trunk region. Taken together, we show that PIs used in combination with platinum agents or topoisomerase inhibitors exhibit increased efficiency with almost 100% inhibition in TNBC cell lines, indicating that PIs are therefore promising compounds to use as combination therapy for TNBC.
© 2024. The Author(s).
Conflict of interest statement
HZ has served at scientific advisory boards and/or as a consultant for Abbvie, Acumen, Alector, Alzinova, ALZPath, Annexon, Apellis, Artery Therapeutics, AZTherapies, Cognito Therapeutics, CogRx, Denali, Eisai, Merry Life, Nervgen, Novo Nordisk, Optoceutics, Passage Bio, Pinteon Therapeutics, Prothena, Red Abbey Labs, reMYND, Roche, Samumed, Siemens Healthineers, Triplet Therapeutics, and Wave, has given lectures in symposia sponsored by Alzecure, Biogen, Cellectricon, Fujirebio, Lilly, and Roche, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program (outside submitted work). The other authors declare no competing interests.
Figures


Similar articles
-
CIP2A is a target of bortezomib in human triple negative breast cancer cells.Breast Cancer Res. 2012 Apr 26;14(2):R68. doi: 10.1186/bcr3175. Breast Cancer Res. 2012. PMID: 22537901 Free PMC article.
-
A targeted RNAi screen of the breast cancer genome identifies KIF14 and TLN1 as genes that modulate docetaxel chemosensitivity in triple-negative breast cancer.Clin Cancer Res. 2013 Apr 15;19(8):2061-70. doi: 10.1158/1078-0432.CCR-13-0082. Epub 2013 Mar 11. Clin Cancer Res. 2013. PMID: 23479679 Free PMC article.
-
Receptors for luteinizing hormone-releasing hormone (GnRH) as therapeutic targets in triple negative breast cancers (TNBC).Target Oncol. 2015 Sep;10(3):365-73. doi: 10.1007/s11523-014-0340-y. Epub 2014 Oct 9. Target Oncol. 2015. PMID: 25293576
-
Calycosin inhibits triple-negative breast cancer progression through down-regulation of the novel estrogen receptor-α splice variant ER-α30-mediated PI3K/AKT signaling pathway.Phytomedicine. 2023 Sep;118:154924. doi: 10.1016/j.phymed.2023.154924. Epub 2023 Jun 14. Phytomedicine. 2023. PMID: 37393829
-
Combination Treatment with EGFR Inhibitor and Doxorubicin Synergistically Inhibits Proliferation of MCF-7 Cells and MDA-MB-231 Triple-Negative Breast Cancer Cells In Vitro.Int J Mol Sci. 2024 Mar 6;25(5):3066. doi: 10.3390/ijms25053066. Int J Mol Sci. 2024. PMID: 38474312 Free PMC article.
Cited by
-
HIV-protease inhibitors potentiate the activity of carfilzomib in triple-negative breast cancer.Br J Cancer. 2024 Sep;131(5):918-930. doi: 10.1038/s41416-024-02774-9. Epub 2024 Jul 5. Br J Cancer. 2024. PMID: 38969867 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

