Aneuploidy and complex genomic rearrangements in cancer evolution
- PMID: 38286829
- PMCID: PMC7616040
- DOI: 10.1038/s43018-023-00711-y
Aneuploidy and complex genomic rearrangements in cancer evolution
Abstract
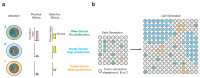
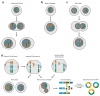
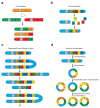
Mutational processes that alter large genomic regions occur frequently in developing tumors. They range from simple copy number gains and losses to the shattering and reassembly of entire chromosomes. These catastrophic events, such as chromothripsis, chromoplexy and the formation of extrachromosomal DNA, affect the expression of many genes and therefore have a substantial effect on the fitness of the cells in which they arise. In this review, we cover large genomic alterations, the mechanisms that cause them and their effect on tumor development and evolution.
© 2024. Springer Nature America, Inc.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Boveri and beyond: Chromothripsis and genomic instability from mitotic errors.Mol Cell. 2024 Jan 4;84(1):55-69. doi: 10.1016/j.molcel.2023.11.002. Epub 2023 Nov 28. Mol Cell. 2024. PMID: 38029753 Review.
-
Prevalence and clinical implications of chromothripsis in cancer genomes.Curr Opin Oncol. 2014 Jan;26(1):64-72. doi: 10.1097/CCO.0000000000000038. Curr Opin Oncol. 2014. PMID: 24305569 Review.
-
The Genomic Characteristics and Origin of Chromothripsis.Methods Mol Biol. 2018;1769:3-19. doi: 10.1007/978-1-4939-7780-2_1. Methods Mol Biol. 2018. PMID: 29564814 Review.
-
Chromothripsis and cancer: causes and consequences of chromosome shattering.Nat Rev Cancer. 2012 Oct;12(10):663-70. doi: 10.1038/nrc3352. Epub 2012 Sep 13. Nat Rev Cancer. 2012. PMID: 22972457 Review.
-
Mechanistic origins of diverse genome rearrangements in cancer.Semin Cell Dev Biol. 2022 Mar;123:100-109. doi: 10.1016/j.semcdb.2021.03.003. Epub 2021 Apr 3. Semin Cell Dev Biol. 2022. PMID: 33824062 Free PMC article. Review.
Cited by
-
CENP-C-Mis12 complex establishes a regulatory loop through Aurora B for chromosome segregation.Life Sci Alliance. 2024 Oct 21;8(1):e202402927. doi: 10.26508/lsa.202402927. Print 2025 Jan. Life Sci Alliance. 2024. PMID: 39433344 Free PMC article.
-
The History of Chromosomal Instability in Genome-Doubled Tumors.Cancer Discov. 2024 Oct 4;14(10):1810-1822. doi: 10.1158/2159-8290.CD-23-1249. Cancer Discov. 2024. PMID: 38943574 Free PMC article.
-
Chromothripsis is rare in IDH-mutant gliomas compared to IDH-wild-type glioblastomas whereas whole-genome duplication is equally frequent in both tumor types.Neurooncol Adv. 2024 Apr 18;6(1):vdae059. doi: 10.1093/noajnl/vdae059. eCollection 2024 Jan-Dec. Neurooncol Adv. 2024. PMID: 38800696 Free PMC article.
-
Dynamics of karyotype evolution.Chaos. 2024 May 1;34(5):051502. doi: 10.1063/5.0206011. Chaos. 2024. PMID: 38717409
-
Loss of heterozygosity impacts MHC expression on the immune microenvironment in CDK12-mutated prostate cancer.Mol Cytogenet. 2024 May 4;17(1):11. doi: 10.1186/s13039-024-00680-6. Mol Cytogenet. 2024. PMID: 38704603 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

