The tRNA thiolation-mediated translational control is essential for plant immunity
- PMID: 38284752
- PMCID: PMC10863982
- DOI: 10.7554/eLife.93517
The tRNA thiolation-mediated translational control is essential for plant immunity
Abstract
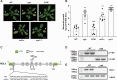
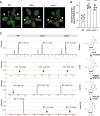
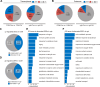
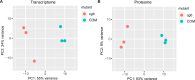
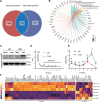
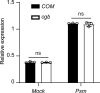
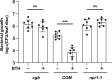
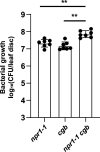
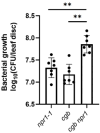
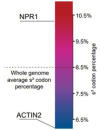
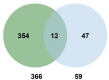
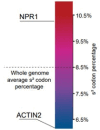
Plants have evolved sophisticated mechanisms to regulate gene expression to activate immune responses against pathogen infections. However, how the translation system contributes to plant immunity is largely unknown. The evolutionarily conserved thiolation modification of transfer RNA (tRNA) ensures efficient decoding during translation. Here, we show that tRNA thiolation is required for plant immunity in Arabidopsis. We identify a cgb mutant that is hyper-susceptible to the pathogen Pseudomonas syringae. CGB encodes ROL5, a homolog of yeast NCS6 required for tRNA thiolation. ROL5 physically interacts with CTU2, a homolog of yeast NCS2. Mutations in either ROL5 or CTU2 result in loss of tRNA thiolation. Further analyses reveal that both transcriptome and proteome reprogramming during immune responses are compromised in cgb. Notably, the translation of salicylic acid receptor NPR1 is reduced in cgb, resulting in compromised salicylic acid signaling. Our study not only reveals a regulatory mechanism for plant immunity but also uncovers an additional biological function of tRNA thiolation.
Keywords: A. thaliana; Arabidopsis; NPR1; plant biology; plant immunity; tRNA thiolation; translation.
© 2024, Zheng, Chen et al.
Conflict of interest statement
XZ, HC, ZD, YW, LZ, CW, XY, QC, SY No competing interests declared
Figures

Update of
- doi: 10.1101/2022.02.13.480182
Similar articles
-
The cytosolic thiouridylase CTU2 of Arabidopsis thaliana is essential for posttranscriptional thiolation of tRNAs and influences root development.BMC Plant Biol. 2014 Apr 28;14:109. doi: 10.1186/1471-2229-14-109. BMC Plant Biol. 2014. PMID: 24774365 Free PMC article.
-
Loss of a Conserved tRNA Anticodon Modification Perturbs Plant Immunity.PLoS Genet. 2015 Oct 22;11(10):e1005586. doi: 10.1371/journal.pgen.1005586. eCollection 2015 Oct. PLoS Genet. 2015. Retraction in: PLoS Genet. 2017 Apr 24;13(4):e1006747. doi: 10.1371/journal.pgen.1006747. PMID: 26492405 Free PMC article. Retracted.
-
Functional analysis of the Theobroma cacao NPR1 gene in Arabidopsis.BMC Plant Biol. 2010 Nov 15;10:248. doi: 10.1186/1471-2229-10-248. BMC Plant Biol. 2010. PMID: 21078185 Free PMC article.
-
NPR1 in JazzSet with Pathogen Effectors.Trends Plant Sci. 2018 Jun;23(6):469-472. doi: 10.1016/j.tplants.2018.04.007. Epub 2018 May 9. Trends Plant Sci. 2018. PMID: 29753632 Review.
-
Research Progress of ATGs Involved in Plant Immunity and NPR1 Metabolism.Int J Mol Sci. 2021 Nov 9;22(22):12093. doi: 10.3390/ijms222212093. Int J Mol Sci. 2021. PMID: 34829975 Free PMC article. Review.
Cited by
-
RNA-Seq Bulked Segregant Analysis of an Exotic B. napus ssp. napobrassica (Rutabaga) F2 Population Reveals Novel QTLs for Breeding Clubroot-Resistant Canola.Int J Mol Sci. 2024 Apr 23;25(9):4596. doi: 10.3390/ijms25094596. Int J Mol Sci. 2024. PMID: 38731814 Free PMC article.
References
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

