Interactions of plumbagin with five common antibiotics against Staphylococcus aureus in vitro
- PMID: 38277418
- PMCID: PMC10817181
- DOI: 10.1371/journal.pone.0297493
Interactions of plumbagin with five common antibiotics against Staphylococcus aureus in vitro
Abstract
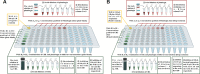
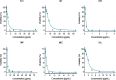
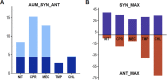
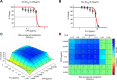
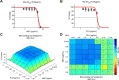
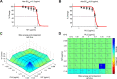
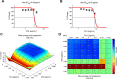
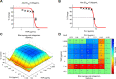
Staphylococcus aureus is the main culprit, causing a variety of severe clinical infections. At the same time, clinics are also facing the severe situation of antibiotic resistance. Therefore, effective strategies to address this problem may include expanding the antimicrobial spectrum by exploring alternative sources of drugs or delaying the development of antibiotic resistance through combination therapy so that existing antibiotics can continue to be used. Plumbagin (PLU) is a phytochemical that exhibits antibacterial activity. In the present study, we investigated the in vitro antibacterial activity of PLU. We selected five antibiotics with different mechanisms and inhibitory activities against S. aureus to explore their interaction with the combination of PLU. The interaction of combinations was evaluated by the Bliss independent model and visualized through response surface analysis. PLU exhibited potent antibacterial activity, with half maximal inhibitory concentration (IC50) and minimum inhibitory concentration (MIC) values against S. aureus of 1.73 μg/mL and 4 μg/mL, respectively. Synergism was observed when PLU was combined with nitrofurantoin (NIT), ciprofloxacin (CPR), mecillinam (MEC), and chloramphenicol (CHL). The indifference of the trimethoprim (TMP)-PLU pairing was demonstrated across the entire dose-response matrix, but significant synergy was observed within a specific dose region. In addition, no antagonistic interactions were indicated. Overall, PLU is not only a promising antimicrobial agent but also has the potential to enhance the growth-inhibitory activity of some antibiotics against S. aureus, and the use of the interaction landscape, along with the dose-response matrix, for analyzing and quantifying combination results represents an improved approach to comprehending antibacterial combinations.
Copyright: © 2024 Bie et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
In vitro antibacterial activity of plumbagin isolated from Plumbago zeylanica L. against methicillin-resistant Staphylococcus aureus.Lett Appl Microbiol. 2019 Jul;69(1):41-49. doi: 10.1111/lam.13160. Epub 2019 May 2. Lett Appl Microbiol. 2019. PMID: 31044446
-
Synergistic Antibacterial Activity Between 1,4-Naphthoquinone and β-Lactam Antibiotics Against Methicillin-Resistant Staphylococcus aureus.Microb Drug Resist. 2021 Feb;27(2):234-240. doi: 10.1089/mdr.2020.0178. Epub 2020 Jun 23. Microb Drug Resist. 2021. PMID: 32589487
-
Synergistic effect of non-steroidal anti-inflammatory drugs (NSAIDs) on antibacterial activity of cefuroxime and chloramphenicol against methicillin-resistant Staphylococcus aureus.J Glob Antimicrob Resist. 2017 Sep;10:70-74. doi: 10.1016/j.jgar.2017.03.012. Epub 2017 Jun 30. J Glob Antimicrob Resist. 2017. PMID: 28673701
-
The Antibacterial Potential of Ciprofloxacin Hybrids against Staphylococcus aureus.Curr Top Med Chem. 2022;22(12):1020-1034. doi: 10.2174/1568026622666220317162132. Curr Top Med Chem. 2022. PMID: 35301951 Review.
-
Staph wars: the antibiotic pipeline strikes back.Microbiology (Reading). 2023 Sep;169(9):001387. doi: 10.1099/mic.0.001387. Microbiology (Reading). 2023. PMID: 37656158 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

