Norovirus Epidemiology and Genotype Circulation during the COVID-19 Pandemic in Brazil, 2019-2022
- PMID: 38276149
- PMCID: PMC10818385
- DOI: 10.3390/pathogens13010003
Norovirus Epidemiology and Genotype Circulation during the COVID-19 Pandemic in Brazil, 2019-2022
Abstract
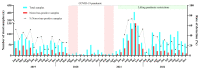
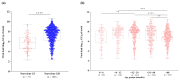
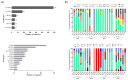
Norovirus stands out as a leading cause of acute gastroenteritis (AGE) worldwide, affecting all age groups. In the present study, we investigated fecal samples from medically attended AGE patients received from nine Brazilian states, from 2019 to 2022, including the COVID-19 pandemic period. Norovirus GI and GII were detected and quantified using RT-qPCR, and norovirus-positive samples underwent genotyping through sequencing the ORF1/2 junction region. During the four-year period, norovirus prevalence was 37.2%, varying from 20.1% in 2020 to 55.4% in 2021. GII genotypes dominated, being detected in 92.9% of samples. GII-infected patients had significantly higher viral concentrations compared to GI-infected patients (median of 3.8 × 107 GC/g and 6.7 × 105 GC/g, respectively); and patients aged >12-24 months showed a higher median viral load (8 × 107 GC/g) compared to other age groups. Norovirus sequencing revealed 20 genotypes by phylogenetic analysis of RdRp and VP1 partial regions. GII.4 Sydney[P16] was the dominant genotype (57.3%), especially in 2019 and 2021, followed by GII.2[P16] (14.8%) and GII.6[P7] (6.3%). The intergenogroup recombinant genotype, GIX.1[GII.P15], was detected in five samples. Our study is the first to explore norovirus epidemiology and genotype distribution in Brazil during COVID-19, and contributes to understanding the epidemiological dynamics of norovirus and highlighting the importance of continuing to follow norovirus surveillance programs in Brazil.
Keywords: Brazil; RT-qPCR; genotyping; molecular epidemiology; norovirus; viral load.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Virological and Epidemiological Features of Norovirus Infections in Brazil, 2017-2018.Viruses. 2021 Aug 30;13(9):1724. doi: 10.3390/v13091724. Viruses. 2021. PMID: 34578304 Free PMC article.
-
Recombinant noroviruses detected in Mid-West region of Brazil in two different periods 2009-2011 and 2014-2015: Atypical breakpoints of recombination and detection of distinct GII.P7-GII.6 lineages.Infect Genet Evol. 2019 Mar;68:47-53. doi: 10.1016/j.meegid.2018.12.007. Epub 2018 Dec 5. Infect Genet Evol. 2019. PMID: 30529559
-
Epidemiological, Molecular, and Clinical Features of Norovirus Infections among Pediatric Patients in Qatar.Viruses. 2019 Apr 29;11(5):400. doi: 10.3390/v11050400. Viruses. 2019. PMID: 31035642 Free PMC article.
-
Molecular epidemiology and genetic diversity of norovirus infection in children hospitalized with acute gastroenteritis in East Java, Indonesia in 2015-2019.Infect Genet Evol. 2021 Mar;88:104703. doi: 10.1016/j.meegid.2020.104703. Epub 2021 Jan 2. Infect Genet Evol. 2021. PMID: 33401005
-
Molecular epidemiology of noroviruses associated with acute sporadic gastroenteritis in children: global distribution of genogroups, genotypes and GII.4 variants.J Clin Virol. 2013 Mar;56(3):185-93. doi: 10.1016/j.jcv.2012.11.011. Epub 2012 Dec 5. J Clin Virol. 2013. PMID: 23218993 Review.
Cited by
-
Genetic Diversity and Phylogenetic Relationship of Human Norovirus Sequences Derived from Municipalities within the Sverdlovsk Region of Russia.Viruses. 2024 Jun 21;16(7):1001. doi: 10.3390/v16071001. Viruses. 2024. PMID: 39066164 Free PMC article.
-
Human norovirus in Brazil: an update of reports in different settings.Braz J Microbiol. 2024 Sep;55(3):2767-2782. doi: 10.1007/s42770-024-01444-5. Epub 2024 Jul 16. Braz J Microbiol. 2024. PMID: 39012425 Review.
-
Diverse genotypes of norovirus genogroup I and II contamination in environmental water in Thailand during the COVID-19 outbreak from 2020 to 2022.Virol Sin. 2024 Aug;39(4):556-564. doi: 10.1016/j.virs.2024.05.010. Epub 2024 May 30. Virol Sin. 2024. PMID: 38823781 Free PMC article.
References
-
- Pires S.M., Fischer-Walker C.L., Lanata C.F., Devleesschauwer B., Hall A.J., Kirk M.D., Duarte A.S.R., Black R.E., Angulo F.J. Aetiology-Specific Estimates of the Global and Regional Incidence and Mortality of Diarrhoeal Diseases Commonly Transmitted through Food. PLoS ONE. 2015;10:e0142927. doi: 10.1371/journal.pone.0142927. - DOI - PMC - PubMed
-
- Lanata C.F., Fischer-Walker C.L., Olascoaga A.C., Torres C.X., Aryee M.J., Black R.E., for the Child Health Epidemiology Reference Group of the World Health Organization and UNICEF Global Causes of Diarrheal Disease Mortality in Children <5 Years of Age: A Systematic Review. PLoS ONE. 2013;8:e72788. doi: 10.1371/journal.pone.0072788. - DOI - PMC - PubMed
-
- Lopman B. Global Burden of Norovirus and Prospects for Vaccine Development. CDC Foundation; Atlanta, GA, USA: 2015. 46p
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

