Deciphering the Role of ERBB3 Isoforms in Renal Cell Carcinoma: A Comprehensive Genomic and Transcriptomic Analysis
- PMID: 38276060
- PMCID: PMC10820170
- DOI: 10.3390/medicina60010181
Deciphering the Role of ERBB3 Isoforms in Renal Cell Carcinoma: A Comprehensive Genomic and Transcriptomic Analysis
Abstract
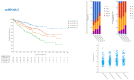
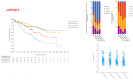
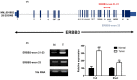
ERBB3, a key member of the receptor tyrosine kinase family, is implicated in the progression and development of various human cancers, affecting cellular proliferation and survival. This study investigated the expression of ERBB3 isoforms in renal clear cell carcinoma (RCC), utilizing data from 538 patients from The Cancer Genome Atlas (TCGA) Firehose Legacy dataset. Employing the SUPPA2 tool, the activity of 10 ERBB3 isoforms was examined, revealing distinct expression patterns in RCC. Isoforms uc001sjg.3 and uc001sjh.3 were found to have reduced activity in tumor tissues, while uc010sqb.2 and uc001sjl.3 demonstrated increased activity. These variations in isoform expression correlate with patient survival and tumor aggressiveness, indicating their complex role in RCC. The study, further, utilizes CIBERSORTx to analyze the association between ERBB3 isoforms and immune cell profiles in the tumor microenvironment. Concurrently, Gene Set Enrichment Analysis (GSEA) was applied, establishing a strong link between elevated levels of ERBB3 isoforms and critical oncogenic pathways, including DNA repair and androgen response. RT-PCR analysis targeting the exon 21-23 and exon 23 regions of ERBB3 confirmed its heightened expression in tumor tissues, underscoring the significance of alternative splicing and exon utilization in cancer development. These findings elucidate the diverse impacts of ERBB3 isoforms on RCC, suggesting their potential as diagnostic markers and therapeutic targets. This study emphasizes the need for further exploration into the specific roles of these isoforms, which could inform more personalized and effective treatment modalities for renal clear cell carcinoma.
Keywords: ERBB3 isoforms; TCGA; renal cell carcinoma.
Conflict of interest statement
The authors have no conflicts of interest.
Figures


Similar articles
-
Comprehensive characterization of alternative splicing in renal cell carcinoma.Brief Bioinform. 2021 Sep 2;22(5):bbab084. doi: 10.1093/bib/bbab084. Brief Bioinform. 2021. PMID: 33822848
-
Comprehensive Multi-Omics Identification of Interferon-γ Response Characteristics Reveals That RBCK1 Regulates the Immunosuppressive Microenvironment of Renal Cell Carcinoma.Front Immunol. 2021 Nov 2;12:734646. doi: 10.3389/fimmu.2021.734646. eCollection 2021. Front Immunol. 2021. PMID: 34795663 Free PMC article.
-
Transcriptomic signatures related to the obesity paradox in patients with clear cell renal cell carcinoma: a cohort study.Lancet Oncol. 2020 Feb;21(2):283-293. doi: 10.1016/S1470-2045(19)30797-1. Epub 2019 Dec 20. Lancet Oncol. 2020. PMID: 31870811 Free PMC article.
-
The ERBB3 receptor in cancer and cancer gene therapy.Cancer Gene Ther. 2008 Jul;15(7):413-48. doi: 10.1038/cgt.2008.15. Epub 2008 Apr 11. Cancer Gene Ther. 2008. PMID: 18404164 Free PMC article. Review.
-
Targeting of erbB3 receptor to overcome resistance in cancer treatment.Mol Cancer. 2014 May 8;13:105. doi: 10.1186/1476-4598-13-105. Mol Cancer. 2014. PMID: 24886126 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

