Rapamycin as a Potential Alternative Drug for Squamous Cell Gingiva Carcinoma (Ca9-22): A Focus on Cell Cycle, Apoptosis and Autophagy Genetic Profile
- PMID: 38276004
- PMCID: PMC10818555
- DOI: 10.3390/ph17010131
Rapamycin as a Potential Alternative Drug for Squamous Cell Gingiva Carcinoma (Ca9-22): A Focus on Cell Cycle, Apoptosis and Autophagy Genetic Profile
Abstract
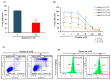
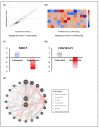
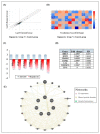
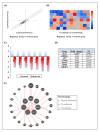
Oral cancer is considered as one of the most common malignancies worldwide. Its conventional treatment primarily involves surgery with or without postoperative adjuvant therapy. The targeting of signaling pathways implicated in tumorigenesis is becoming increasingly prevalent in the development of new anticancer drug candidates. Based on our recently published data, Rapamycin, an inhibitor of the mTOR pathway, exhibits selective antitumor activity in oral cancer by inhibiting cell proliferation and inducing cancer cell apoptosis, autophagy, and cellular stress. In the present study, our focus is on elucidating the genetic determinants of Rapamycin's action and the interaction networks accountable for tumorigenesis suppression. To achieve this, gingival carcinoma cell lines (Ca9-22) were exposed to Rapamycin at IC50 (10 µM) for 24 h. Subsequently, we investigated the genetic profiles related to the cell cycle, apoptosis, and autophagy, as well as gene-gene interactions, using QPCR arrays and the Gene MANIA website. Overall, our results showed that Rapamycin at 10 µM significantly inhibits the growth of Ca9-22 cells after 24 h of treatment by around 50% by suppression of key modulators in the G2/M transition, namely, Survivin and CDK5RAP1. The combination of Rapamycin with Cisplatin potentializes the inhibition of Ca9-22 cell proliferation. A P1/Annexin-V assay was performed to evaluate the effect of Rapamycin on cell apoptosis. The results obtained confirm our previous findings in which Rapamycin at 10 μM induces a strong apoptosis of Ca9-22 cells. The live cells decreased, and the late apoptotic cells increased when the cells were treated by Rapamycin. To identify the genes responsible for cell apoptosis induced by Rapamycin, we performed the RT2 Profiler PCR Arrays for 84 apoptotic genes. The blocked cells were believed to be directed towards cell death, confirmed by the downregulation of apoptosis inhibitors involved in both the extrinsic and intrinsic pathways, including BIRC5, BNIP3, CD40LG, DAPK1, LTA, TNFRSF21 and TP73. The observed effects of Rapamycin on tumor suppression are likely to involve the autophagy process, evidenced by the inhibition of autophagy modulators (TGFβ1, RGS19 and AKT1), autophagosome biogenesis components (AMBRA1, ATG9B and TMEM74) and autophagy byproducts (APP). Identifying gene-gene interaction (GGI) networks provided a comprehensive view of the drug's mechanism and connected the studied tumorigenesis processes to potential functional interactions of various kinds (physical interaction, co-expression, genetic interactions etc.). In conclusion, Rapamycin shows promise as a clinical agent for managing Ca9-22 gingiva carcinoma cells.
Keywords: Rapamycin; apoptosis; autophagy; cell cycle; genetic profile; oral cancer.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
mTOR is a fine tuning molecule in CDK inhibitors-induced distinct cell death mechanisms via PI3K/AKT/mTOR signaling axis in prostate cancer cells.Apoptosis. 2016 Oct;21(10):1158-78. doi: 10.1007/s10495-016-1275-9. Apoptosis. 2016. PMID: 27484210
-
Rapamycin inhibits lung squamous cell carcinoma growth by downregulating glypican-3/Wnt/β-catenin signaling and autophagy.J Cancer Res Clin Oncol. 2021 Feb;147(2):499-505. doi: 10.1007/s00432-020-03422-4. Epub 2020 Nov 23. J Cancer Res Clin Oncol. 2021. PMID: 33225417
-
ABT-751 Induces Multiple Anticancer Effects in Urinary Bladder Urothelial Carcinoma-Derived Cells: Highlighting the Induction of Cytostasis through the Inhibition of SKP2 at Both Transcriptional and Post-Translational Levels.Int J Mol Sci. 2021 Jan 19;22(2):945. doi: 10.3390/ijms22020945. Int J Mol Sci. 2021. PMID: 33478005 Free PMC article.
-
Synergistic Effects of New Curcumin Analog (PAC) and Cisplatin on Oral Cancer Therapy.Curr Issues Mol Biol. 2023 Jun 8;45(6):5018-5035. doi: 10.3390/cimb45060319. Curr Issues Mol Biol. 2023. PMID: 37367068 Free PMC article.
-
Rapamycin inhibits oral cancer cell growth by promoting oxidative stress and suppressing ERK1/2, NF-κB and beta-catenin pathways.Front Oncol. 2022 Sep 15;12:873447. doi: 10.3389/fonc.2022.873447. eCollection 2022. Front Oncol. 2022. PMID: 36185289 Free PMC article.
References
-
- World Health Organization . WHO Highlights Oral Health Neglect Affecting Nearly Half of the World’s Population. World Health Organization; Geneva, Switzerland: 2022. Global Oral Health Status Report 2022.
-
- National Cancer Institute . Side Effects of Cancer Treatment. National Cancer Institute; Bethesda, MD, USA: 2021.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

