MAGOH promotes gastric cancer progression via hnRNPA1 expression inhibition-mediated RONΔ160/PI3K/AKT signaling pathway activation
- PMID: 38268030
- PMCID: PMC10809607
- DOI: 10.1186/s13046-024-02946-8
MAGOH promotes gastric cancer progression via hnRNPA1 expression inhibition-mediated RONΔ160/PI3K/AKT signaling pathway activation
Abstract
Background: Gastric cancer (GC) is associated with high mortality and heterogeneity and poses a great threat to humans. Gene therapies for the receptor tyrosine kinase RON and its spliceosomes are attracting increasing amounts of attention due to their unique characteristics. However, little is known about the mechanism involved in the formation of the RON mRNA alternative spliceosome RONΔ160.
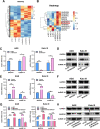
Methods: Fourteen human GC tissue samples and six normal gastric tissue samples were subjected to label-free relative quantitative proteomics analysis, and MAGOH was identified as a candidate protein for subsequent studies. The expression of MAGOH in clinical specimens was verified by quantitative real-time PCR and western blotting. We then determined the biological function of MAGOH in GC through in vitro and in vivo experiments. RNA pulldown, RNA sequencing and RNA immunoprecipitation (RIP) were subsequently conducted to uncover the underlying mechanism by which MAGOH regulated the formation of RONΔ160.
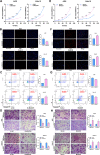
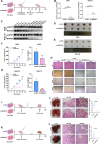
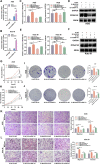
Results: Proteomic analysis revealed that MAGOH, which is located at key nodes and participates in RNA processing and mRNA splicing, was upregulated in GC tissue and GC cell lines and was associated with poor prognosis. Functional analysis showed that MAGOH promoted the proliferation, migration and invasion of GC cells in vitro and in vivo. Mechanistically, MAGOH inhibited the expression of hnRNPA1 and reduced the binding of hnRNPA1 to RON mRNA, thereby promoting the formation of RONΔ160 to activate the PI3K/AKT signaling pathway and consequently facilitating GC progression.
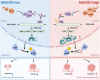
Conclusions: Our study revealed that MAGOH could promote the formation of RONΔ160 and activate the PI3K/AKT signaling pathway through the inhibition of hnRNPA1 expression. We elucidate a novel mechanism and potential therapeutic targets for the growth and metastasis of GC based on the MAGOH-RONΔ160 axis, and these findings have important guiding significance and clinical value for the future development of effective therapeutic strategies for GC.
Keywords: Alternative splicing; Gastric cancer; MAGOH; PI3K/AKT signaling pathway; RONΔ160; hnRNPA1.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest. Figure 9 was created with BioRender software.
Figures



Similar articles
-
HOXB13 promotes gastric cancer cell migration and invasion via IGF-1R upregulation and subsequent activation of PI3K/AKT/mTOR signaling pathway.Life Sci. 2021 Aug 1;278:119522. doi: 10.1016/j.lfs.2021.119522. Epub 2021 Apr 21. Life Sci. 2021. PMID: 33894267
-
RON and RONΔ160 promote gastric cancer cell proliferation, migration, and adaption to hypoxia via interaction with β-catenin.Aging (Albany NY). 2019 May 13;11(9):2735-2748. doi: 10.18632/aging.101945. Aging (Albany NY). 2019. PMID: 31085796 Free PMC article.
-
Mir-20a-5p induced WTX deficiency promotes gastric cancer progressions through regulating PI3K/AKT signaling pathway.J Exp Clin Cancer Res. 2020 Oct 8;39(1):212. doi: 10.1186/s13046-020-01718-4. J Exp Clin Cancer Res. 2020. PMID: 33032635 Free PMC article.
-
Targeting of SPP1 by microRNA-340 inhibits gastric cancer cell epithelial-mesenchymal transition through inhibition of the PI3K/AKT signaling pathway.J Cell Physiol. 2019 Aug;234(10):18587-18601. doi: 10.1002/jcp.28497. Epub 2019 Apr 5. J Cell Physiol. 2019. Retraction in: J Cell Physiol. 2022 Mar;237(3):2010. doi: 10.1002/jcp.30522 PMID: 30953349 Retracted.
-
ACTL8 Promotes the Progression of Gastric Cancer Through PI3K/AKT/mTOR Signaling Pathway.Dig Dis Sci. 2024 Oct;69(10):3786-3798. doi: 10.1007/s10620-024-08649-6. Epub 2024 Sep 25. Dig Dis Sci. 2024. PMID: 39322809 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

