Phylogeny and diversification of genus Sanicula L. (Apiaceae): novel insights from plastid phylogenomic analyses
- PMID: 38263006
- PMCID: PMC10807117
- DOI: 10.1186/s12870-024-04750-0
Phylogeny and diversification of genus Sanicula L. (Apiaceae): novel insights from plastid phylogenomic analyses
Abstract
Background: The genus Sanicula L. is a unique perennial herb that holds important medicinal values. Although the previous studies on Sanicula provided us with a good research basis, its taxonomic system and interspecific relationships have not been satisfactorily resolved, especially for those endemic to China. Moreover, the evolutionary history of this genus also remains inadequately understood. The plastid genomes possessing highly conserved structure and limited evolutionary rate have proved to be an effective tool for studying plant phylogeny and evolution.
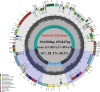
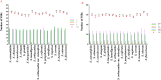
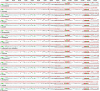
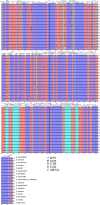
Results: In the current study, we newly sequenced and assembled fifteen Sanicula complete plastomes. Combined with two previously reported plastomes, we performed comprehensively plastid phylogenomics analyses to gain novel insights into the evolutionary history of this genus. The comparative results indicated that the seventeen plastomes exhibited a high degree of conservation and similarity in terms of their structure, size, GC content, gene order, IR borders, codon bias patterns and SSRs profiles. Such as all of them displayed a typical quadripartite structure, including a large single copy region (LSC: 85,074-86,197 bp), a small single copy region (SSC: 17,047-17,132 bp) separated by a pair of inverted repeat regions (IRs: 26,176-26,334 bp). And the seventeen plastomes had similar IR boundaries and the adjacent genes were identical. The rps19 gene was located at the junction of the LSC/IRa, the IRa/SSC junction region was located between the trnN gene and ndhF gene, the ycf1 gene appeared in the SSC/IRb junction and the IRb/LSC boundary was located between rpl12 gene and trnH gene. Twelve specific mutation hotspots (atpF, cemA, accD, rpl22, rbcL, matK, ycf1, trnH-psbA, ycf4-cemA, rbcL-accD, trnE-trnT and trnG-trnR) were identified that can serve as potential DNA barcodes for species identification within the genus Sanicula. Furthermore, the plastomes data and Internal Transcribed Spacer (ITS) sequences were performed to reconstruct the phylogeny of Sanicula. Although the tree topologies of them were incongruent, both provided strong evidence supporting the monophyly of Saniculoideae and Apioideae. In addition, the sister groups between Saniculoideae and Apioideae were strongly suggested. The Sanicula species involved in this study were clustered into a clade, and the Eryngium species were also clustered together. However, it was clearly observed that the sections of Sanicula involved in the current study were not respectively recovered as monophyletic group. Molecular dating analysis explored that the origin of this genus was occurred during the late Eocene period, approximately 37.84 Ma (95% HPD: 20.33-52.21 Ma) years ago and the diversification of the genus was occurred in early Miocene 18.38 Ma (95% HPD: 10.68-25.28 Ma).
Conclusion: The plastome-based tree and ITS-based tree generated incongruences, which may be attributed to the event of hybridization/introgression, incomplete lineage sorting (ILS) and chloroplast capture. Our study highlighted the power of plastome data to significantly improve the phylogenetic supports and resolutions, and to efficiently explore the evolutionary history of this genus. Molecular dating analysis explored that the diversification of the genus occurred in the early Miocene, which was largely influenced by the prevalence of the East Asian monsoon and the uplift of the Hengduan Mountains (HDM). In summary, our study provides novel insights into the plastome evolution, phylogenetic relationships, taxonomic framework and evolution of genus Sanicula.
Keywords: Apiaceae; DNA barcodes; Divergence time; Phylogeny; Plastome; Sanicula.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Complete chloroplast of four Sanicula taxa (Apiaceae) endemic to China: lights into genome structure, comparative analysis, and phylogenetic relationships.BMC Plant Biol. 2023 Sep 21;23(1):444. doi: 10.1186/s12870-023-04447-w. BMC Plant Biol. 2023. PMID: 37730528 Free PMC article.
-
Characteristics of plastid genomes in the genus Ceratostigma inhabiting arid habitats in China and their phylogenomic implications.BMC Plant Biol. 2023 Jun 7;23(1):303. doi: 10.1186/s12870-023-04323-7. BMC Plant Biol. 2023. PMID: 37280518 Free PMC article.
-
The complete plastomes of thirteen Libanotis (Apiaceae, Apioideae) plants: comparative and phylogenetic analyses provide insights into the plastome evolution and taxonomy of Libanotis.BMC Plant Biol. 2024 Feb 12;24(1):106. doi: 10.1186/s12870-024-04784-4. BMC Plant Biol. 2024. PMID: 38342898 Free PMC article.
-
Dynamic evolution of the plastome in the Elm family (Ulmaceae).Planta. 2022 Dec 17;257(1):14. doi: 10.1007/s00425-022-04045-4. Planta. 2022. PMID: 36526857 Review.
-
A reference cell tree will serve science better than a reference cell atlas.Cell. 2023 Mar 16;186(6):1103-1114. doi: 10.1016/j.cell.2023.02.016. Cell. 2023. PMID: 36931241 Review.
Cited by
-
The Micromorphology and Its Taxonomic Value of the Genus Sanicula L. in China (Apiaceae).Plants (Basel). 2024 Jun 13;13(12):1635. doi: 10.3390/plants13121635. Plants (Basel). 2024. PMID: 38931068 Free PMC article.
-
Phylogeny, adaptive evolution, and taxonomy of Acronema (Apiaceae): evidence from plastid phylogenomics and morphological data.Front Plant Sci. 2024 Aug 16;15:1425158. doi: 10.3389/fpls.2024.1425158. eCollection 2024. Front Plant Sci. 2024. PMID: 39220016 Free PMC article.
-
Plastid phylogenomics contributes to the taxonomic revision of taxa within the genus Sanicula L. and acceptance of two new members of the genus.Front Plant Sci. 2024 Jun 10;15:1351023. doi: 10.3389/fpls.2024.1351023. eCollection 2024. Front Plant Sci. 2024. PMID: 38916035 Free PMC article.
References
-
- Pryer KM, Phillippe LR. A synopsis of the genus Sanicula (Apiaceae) in eastern Canada. Can J Bot. 1989;67(3):694–707. doi: 10.1139/b89-093. - DOI
-
- Pimenov MG, Leonov MV. The genera of the Umbelliferae: a nomenclator. Kew: Royal Botanic Gardens; 1993.
-
- Van Wyk B-E, Tilney PM, Magee AR. African Apiaceae: a synopsis of the Apiaceae/Umbelliferae of Sub-Saharan Africa and Madagascar/Ben-Erik Van Wyk, Patricia M Tilney & Anthony R Magee. Pretoria: Briza Academic Books; 2013.
-
- Koch WDJ. Generum tribriumque plantarum umbelliferarum nova dispositis. Nova Acta Acad Caes Leop-Carol. 1824;12:55–156.
-
- Vargas P, Baldwin BG, Constance L. A phylogenetic study of Sanicula sect. Sanicoria and S. sect. Sandwicenses (Apiaceae) based on nuclear rDNA and morphological data. Syst Bot. 1999;24(2):228. doi: 10.2307/2419550. - DOI
MeSH terms
Grants and funding
- Project No.: 510101202200376/Survey on the Background Resources of Chengdu Area of Giant Panda National Park
- Project No.: 510101202200376/Survey on the Background Resources of Chengdu Area of Giant Panda National Park
- Project No.: 510101202200376/Survey on the Background Resources of Chengdu Area of Giant Panda National Park
- Project No.: 510101202200376/Survey on the Background Resources of Chengdu Area of Giant Panda National Park
- Project No.: 510101202200376/Survey on the Background Resources of Chengdu Area of Giant Panda National Park
- Project No.: 510101202200376/Survey on the Background Resources of Chengdu Area of Giant Panda National Park
- Project No.: 510101202200376/Survey on the Background Resources of Chengdu Area of Giant Panda National Park
- Project No.: 510101202200376/Survey on the Background Resources of Chengdu Area of Giant Panda National Park
- Project No.: 510101202200376/Survey on the Background Resources of Chengdu Area of Giant Panda National Park
LinkOut - more resources
Full Text Sources
Miscellaneous

