This is a preprint.
Tissue-specific RNA-seq defines genes governing male tail tip morphogenesis in C. elegans
- PMID: 38260477
- PMCID: PMC10802606
- DOI: 10.1101/2024.01.12.575210
Tissue-specific RNA-seq defines genes governing male tail tip morphogenesis in C. elegans
Update in
-
Tissue-specific RNA-seq defines genes governing male tail tip morphogenesis in C. elegans.Development. 2024 Sep 15;151(18):dev202787. doi: 10.1242/dev.202787. Epub 2024 Sep 24. Development. 2024. PMID: 39253748
Abstract
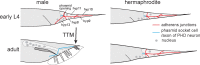
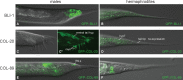
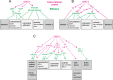
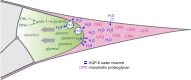
Caenorhabditis elegans males undergo sex-specific tail tip morphogenesis (TTM) under the control of the transcription factor DMD-3. To find genes regulated by DMD-3, We performed RNA-seq of laser-dissected tail tips. We identified 564 genes differentially expressed (DE) in wild-type males vs. dmd-3(-) males and hermaphrodites. The transcription profile of dmd-3(-) tail tips is similar to that in hermaphrodites. For validation, we analyzed transcriptional reporters for 49 genes and found male-specific or male-biased expression for 26 genes. Only 11 DE genes overlapped with genes found in a previous RNAi screen for defective TTM. GO enrichment analysis of DE genes finds upregulation of genes within the UPR (unfolded protein response) pathway and downregulation of genes involved in cuticle maintenance. Of the DE genes, 40 are transcription factors, indicating that the gene network downstream of DMD-3 is complex and potentially modular. We propose modules of genes that act together in TTM and are coregulated by DMD-3, among them the chondroitin synthesis pathway and the hypertonic stress response.
Keywords: DMD-3; GRN; chondroitin proteoglycan; differential expression; gene network.
Figures



Similar articles
-
Tissue-specific RNA-seq defines genes governing male tail tip morphogenesis in C. elegans.Development. 2024 Sep 15;151(18):dev202787. doi: 10.1242/dev.202787. Epub 2024 Sep 24. Development. 2024. PMID: 39253748
-
dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans.Development. 2008 Aug;135(14):2373-82. doi: 10.1242/dev.017046. Epub 2008 Jun 11. Development. 2008. PMID: 18550714 Free PMC article.
-
The novel gene, mtre-1 , is expressed downstream of MAB-3 and DMD-3 in the male tail tip at the termination of male tail tip retraction.MicroPubl Biol. 2023 Oct 18;2023:10.17912/micropub.biology.000976. doi: 10.17912/micropub.biology.000976. eCollection 2023. MicroPubl Biol. 2023. PMID: 37927909 Free PMC article.
-
A bow-tie genetic architecture for morphogenesis suggested by a genome-wide RNAi screen in Caenorhabditis elegans.PLoS Genet. 2011 Mar;7(3):e1002010. doi: 10.1371/journal.pgen.1002010. Epub 2011 Mar 3. PLoS Genet. 2011. PMID: 21408209 Free PMC article.
-
From cell fates to morphology: developmental genetics of the Caenorhabditis elegans male tail.Bioessays. 1992 May;14(5):309-16. doi: 10.1002/bies.950140504. Bioessays. 1992. PMID: 1637362 Review.
References
-
- Asahina M., Thurtle-Schmidt D. and Yamamoto K. R. (2020). Genetic exploration of a nuclear receptor transcriptional regulatory complex. 2020.03.28.013060.
-
- Bernadskaya Y. and Christiaen L. (2016). Transcriptional Control of Developmental Cell Behaviors. Annu. Rev. Cell Dev. Biol. 32, 77–101. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
