Design of protein-binding peptides with controlled binding affinity: the case of SARS-CoV-2 receptor binding domain and angiotensin-converting enzyme 2 derived peptides
- PMID: 38250735
- PMCID: PMC10797010
- DOI: 10.3389/fmolb.2023.1332359
Design of protein-binding peptides with controlled binding affinity: the case of SARS-CoV-2 receptor binding domain and angiotensin-converting enzyme 2 derived peptides
Abstract
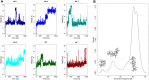
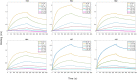
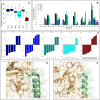
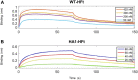
The development of methods able to modulate the binding affinity between proteins and peptides is of paramount biotechnological interest in view of a vast range of applications that imply designed polypeptides capable to impair or favour Protein-Protein Interactions. Here, we applied a peptide design algorithm based on shape complementarity optimization and electrostatic compatibility and provided the first experimental in vitro proof of the efficacy of the design algorithm. Focusing on the interaction between the SARS-CoV-2 Spike Receptor-Binding Domain (RBD) and the human angiotensin-converting enzyme 2 (ACE2) receptor, we extracted a 23-residues long peptide that structurally mimics the major interacting portion of the ACE2 receptor and designed in silico five mutants of such a peptide with a modulated affinity. Remarkably, experimental KD measurements, conducted using biolayer interferometry, matched the in silico predictions. Moreover, we investigated the molecular determinants that govern the variation in binding affinity through molecular dynamics simulation, by identifying the mechanisms driving the different values of binding affinity at a single residue level. Finally, the peptide sequence with the highest affinity, in comparison with the wild type peptide, was expressed as a fusion protein with human H ferritin (HFt) 24-mer. Solution measurements performed on the latter constructs confirmed that peptides still exhibited the expected trend, thereby enhancing their efficacy in RBD binding. Altogether, these results indicate the high potentiality of this general method in developing potent high-affinity vectors for hindering/enhancing protein-protein associations.
Keywords: biolayer interferometry (BLI); ferritin-based nanoparticle; molecular dynamics simulation; peptide design; shape complementarity.
Copyright © 2024 Parisi, Piacentini, Incocciati, Bonamore, Macone, Rupert, Zacco, Miotto, Milanetti, Tartaglia, Ruocco, Boffi and Di Rienzo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


Similar articles
-
Computational Design and Experimental Validation of ACE2-Derived Peptides as SARS-CoV-2 Receptor Binding Domain Inhibitors.J Phys Chem B. 2022 Oct 20;126(41):8129-8139. doi: 10.1021/acs.jpcb.2c03918. Epub 2022 Oct 11. J Phys Chem B. 2022. PMID: 36219223
-
Computational Design of 25-mer Peptide Binders of SARS-CoV-2.J Phys Chem B. 2020 Dec 3;124(48):10930-10942. doi: 10.1021/acs.jpcb.0c07890. Epub 2020 Nov 17. J Phys Chem B. 2020. PMID: 33200935
-
Computational design of SARS-CoV-2 peptide binders with better predicted binding affinities than human ACE2 receptor.Sci Rep. 2021 Aug 2;11(1):15650. doi: 10.1038/s41598-021-94873-3. Sci Rep. 2021. PMID: 34341401 Free PMC article.
-
Shedding Light on the Inhibitory Mechanisms of SARS-CoV-1/CoV-2 Spike Proteins by ACE2-Designed Peptides.J Chem Inf Model. 2021 Mar 22;61(3):1226-1243. doi: 10.1021/acs.jcim.0c01320. Epub 2021 Feb 23. J Chem Inf Model. 2021. PMID: 33619962
-
Electrostatic Interactions Are the Primary Determinant of the Binding Affinity of SARS-CoV-2 Spike RBD to ACE2: A Computational Case Study of Omicron Variants.Int J Mol Sci. 2022 Nov 26;23(23):14796. doi: 10.3390/ijms232314796. Int J Mol Sci. 2022. PMID: 36499120 Free PMC article.
Cited by
-
Self-Assembled Ferritin Nanoparticles for Delivery of Antigens and Development of Vaccines: From Structure and Property to Applications.Molecules. 2024 Sep 5;29(17):4221. doi: 10.3390/molecules29174221. Molecules. 2024. PMID: 39275069 Free PMC article. Review.
-
Computational Approaches to Predict Protein-Protein Interactions in Crowded Cellular Environments.Chem Rev. 2024 Apr 10;124(7):3932-3977. doi: 10.1021/acs.chemrev.3c00550. Epub 2024 Mar 27. Chem Rev. 2024. PMID: 38535831 Free PMC article. Review.
References
-
- Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., et al. (2015). GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25. 10.1016/j.softx.2015.06.001 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

