Identification and validation of methylation-CpG prognostic signature for prognosis of hepatocellular carcinoma
- PMID: 38244582
- PMCID: PMC10866447
- DOI: 10.18632/aging.205454
Identification and validation of methylation-CpG prognostic signature for prognosis of hepatocellular carcinoma
Abstract
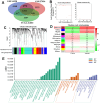
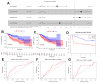
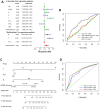
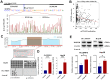
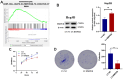
Epigenetic biomarkers help predict the prognosis of cancer patients and evaluating the clinical outcome of immunization therapy. In this study, we present a personalized gene methylation-CpG signature to enhance the accuracy of survival prediction for individuals with hepatocellular carcinoma (HCC). Utilizing RNA sequencing and methylation datasets from GEO as well as TCGA, we conducted single sample GSEA (ssGSEA), WGCNA, as well as Cox regression. Through these analyses, we identified 175 oxidative stress and immune-related genes along with 4 CpG loci that are associated with the prognosis of HCC. Subsequently, we constructed a prognostic signature for HCC utilizing these 4 CpG sites, referred to as the HCC Prognostic Signature of Methylation-CpG sites (HPSM). Further investigation revealed an enrichment of immune-related signal pathways in the HPSM-low group, which demonstrated a positive correlation with better survival among HCC patients. Moreover, the methylation of the CpG sites in HPSM was found to be closely linked to drug sensitivity. In vitro experiments tentatively confirmed that promoter methylation regulated the expression of BMPER, one of the CpG sites within HPSM. The expression of BMPER was significantly correlated with cell death in the oxidative stress pathway, and overexpression of BMPER effectively inhibited HCC cell proliferation. Consequently, our findings suggest that HPSM is an independent predictive factor and holds promise for accurately predicting the prognosis of HCC patients.
Keywords: hepatocellular carcinoma; immune checkpoint; methylation; oxidative stress; prognostic.
Conflict of interest statement
Figures

Similar articles
-
Identification and characterization of a 25-lncRNA prognostic signature for early recurrence in hepatocellular carcinoma.BMC Cancer. 2021 Oct 30;21(1):1165. doi: 10.1186/s12885-021-08827-z. BMC Cancer. 2021. PMID: 34717566 Free PMC article.
-
Characterizing the key genes of COVID-19 that regulate tumor immune microenvironment and prognosis in hepatocellular carcinoma.Funct Integr Genomics. 2023 Aug 4;23(3):262. doi: 10.1007/s10142-023-01184-z. Funct Integr Genomics. 2023. PMID: 37540264
-
Immunological Significance of Prognostic DNA Methylation Sites in Hepatocellular Carcinoma.Front Mol Biosci. 2021 May 26;8:683240. doi: 10.3389/fmolb.2021.683240. eCollection 2021. Front Mol Biosci. 2021. PMID: 34124163 Free PMC article.
-
Comprehensive scRNA-seq Analysis and Identification of CD8_+T Cell Related Gene Markers for Predicting Prognosis and Drug Resistance of Hepatocellular Carcinoma.Curr Med Chem. 2024;31(17):2414-2430. doi: 10.2174/0109298673274578231030065454. Curr Med Chem. 2024. PMID: 37936457
-
Upregulated CANT1 is correlated with poor prognosis in hepatocellular carcinoma.BMC Cancer. 2023 Oct 19;23(1):1007. doi: 10.1186/s12885-023-11463-4. BMC Cancer. 2023. PMID: 37858061 Free PMC article.
References
-
- Lee JH, Suh JH, Choi SY, Kang HJ, Lee HH, Ye BJ, Lee GR, Jung SW, Kim CJ, Lee-Kwon W, Park J, Myung K, Park NH, Kwon HM. Tonicity-responsive enhancer-binding protein promotes hepatocellular carcinogenesis, recurrence and metastasis. Gut. 2019; 68:347–58. 10.1136/gutjnl-2017-315348 - DOI - PMC - PubMed
-
- Fu Y, Silverstein S, McCutcheon JN, Dyba M, Nath RG, Aggarwal M, Coia H, Bai A, Pan J, Jiang J, Kallakury B, Wang H, Zhang YW, et al.. An endogenous DNA adduct as a prognostic biomarker for hepatocarcinogenesis and its prevention by Theaphenon E in mice. Hepatology. 2018; 67:159–70. 10.1002/hep.29380 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

