Genomics and tumor microenvironment of breast mucoepidermoid carcinoma based on whole-exome and RNA sequencing
- PMID: 38243319
- PMCID: PMC10797953
- DOI: 10.1186/s13000-024-01439-8
Genomics and tumor microenvironment of breast mucoepidermoid carcinoma based on whole-exome and RNA sequencing
Abstract
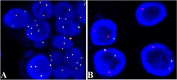
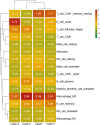
Mammary mucoepidermoid carcinoma (MEC) is a rare entity. The molecular characteristics of breast MEC have not been fully investigated due to its rarity. We performed a retrospective study among 1000 patients with breast carcinomas and identified four cases of breast MEC. Clinical and demographic data were collected. Immunohistochemistry panels which were used to diagnose salivary gland MEC and breast carcinomas were also performed. MAML2 rearrangements were detected by FISH and fusion partners were identified by RNA sequencing. Whole-exome sequencing (WES) was used to reveal the genomes of these four breast MEC. Then, the biological functions and features of breast MEC were further compared with those of invasive breast carcinomas and salivary gland MEC.According to Ellis and Auclair's methods, these four breast MEC could be classified as low-grade breast MEC. All the patients were alive, and disease-free survival (PFS) ranged from 20 months to 67 months. Among these four breast MEC, two cases were triple-negative, and the other two cases were found to be ER positive, with one also showing HER2 equivocal by immunohistochemical staining, but no amplification in FISH. FISH analysis confirmed the presence of the MAML2 translocation in three of four tumors, and CRTC1-MAML2 fusion was confirmed in two of them by RNA-sequencing. The average coverage size of WES for the tumor mutation burden estimation was 32 Mb. MUC4, RP1L1 and QRICH2 mutations were identified in at least three tumors, and these mutation also existed in breast invasive carcinoma databases (TCGA, Cell 2015; TCGA, Nature 2012). The results showed that there were many genes in breast MEC overlapping with the breast invasive carcinoma databases mentioned above, range from 5 to 63 genes (median:21 genes). Next, we assessed immune cell infiltration levels in these tumors. In all these tumors, M2 macrophages and plasma cell were in the high infiltration group. Our breast MEC showed different results from the salivary gland MEC, whose plasma cells were in the low infiltration group. Overall, we first analyzed the genomics and tumor microenvironment of breast mucoepidermoid carcinoma and proposed our hypothesis that although MECs arising in the breast resemble their salivary gland counterparts phenotypically, our findings indicate that breast MECs probably resemble invasive breast carcinomas at the genetic level and immune cell infiltration levels. More cases and in deep research need to be done to further understand this rare carcinoma.
Keywords: Breast; Mucoepidermoid carcinoma; RNA-sequencing; Whole-exome sequencing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
"Pancreatic Mucoepidermoid Carcinoma" Is not a Pancreatic Counterpart of CRTC1/3-MAML2 Fusion Gene-related Mucoepidermoid Carcinoma of the Salivary Gland, and May More Appropriately be Termed Pancreatic Adenosquamous Carcinoma With Mucoepidermoid Carcinoma-like Features.Am J Surg Pathol. 2018 Nov;42(11):1419-1428. doi: 10.1097/PAS.0000000000001135. Am J Surg Pathol. 2018. PMID: 30138216
-
CRTC1-MAML2 fusion in mucoepidermoid carcinoma of the breast.Histopathology. 2019 Feb;74(3):463-473. doi: 10.1111/his.13779. Epub 2018 Dec 5. Histopathology. 2019. PMID: 30380176
-
Reevaluation of MAML2 fusion-negative mucoepidermoid carcinoma: a subgroup being actually hyalinizing clear cell carcinoma of the salivary gland with EWSR1 translocation.Hum Pathol. 2017 Mar;61:9-18. doi: 10.1016/j.humpath.2016.06.029. Epub 2016 Oct 18. Hum Pathol. 2017. PMID: 27769871
-
Mucoepidermoid carcinoma of the esophagus with MAML2 gene rearrangement: Case report and literature review.Pathol Res Pract. 2023 Jan;241:154242. doi: 10.1016/j.prp.2022.154242. Epub 2022 Dec 2. Pathol Res Pract. 2023. PMID: 36481651 Review.
-
Primary mucoepidermoid carcinoma of the external auditory canal with a CRTC1::MAML2 fusion: A case report and a review of literature.J Cutan Pathol. 2023 Nov;50(11):947-950. doi: 10.1111/cup.14491. Epub 2023 Jul 2. J Cutan Pathol. 2023. PMID: 37394842 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

