Decoding the multifaceted interventions between human sirtuin 2 and dynamic hepatitis B viral proteins to confirm their roles in HBV replication
- PMID: 38239506
- PMCID: PMC10794644
- DOI: 10.3389/fcimb.2023.1234903
Decoding the multifaceted interventions between human sirtuin 2 and dynamic hepatitis B viral proteins to confirm their roles in HBV replication
Abstract
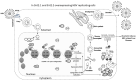
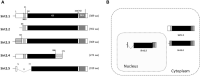
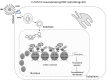
The human sirtuin 2 gene (SIRT2) encodes a full-length Sirt2 protein (i.e., the Sirt2 isoform 1), which primarily functions as a cytoplasmic α-tubulin deacetylase, and which promotes the growth of hepatocellular carcinoma (HCC). Hepatitis B virus (HBV) replication itself, or HBV X (HBx) protein-mediated transcriptional transactivation, enhances Sirt2.1 expression; therefore, Sirt2.1 itself is capable of positively increasing HBV transcription and replication. Sirt2.1 is linked to liver fibrosis and epithelial-to-mesenchymal transition and, consequently, augments the risk of HCC. The Sirt2.1 protein enhances the HBV replication cycle by activating the AKT/glycogen synthase kinase 3 beta (GSK3β)/β-catenin pathway. It also activates the transcription of the viral enhancer I/HBx promoter (EnI/Xp) and enhancer II/HBc promoter (EnII/Cp) by targeting the transcription factor p53. The Sirt2 isoform 2 (Sirt2.2) is mainly localized in the cytoplasm, and the N-terminus is shorter by 37 amino acids than that of Sirt2.1. Despite the truncation of the N-terminal region, Sirt2.2 is still capable of enhancing HBV replication and activating the AKT/GSK3β/β-catenin signaling pathway. The Sirt2 isoform 5 (Sirt2.5) is primarily localized to the nucleus, it lacks a nuclear export signal (NES), and the catalytic domain (CD) is truncated. Upon HBV replication, expression of the Sirt2 isoforms is also enhanced, which further upregulates the HBV replication, and, therefore, supports the vicious cycle of viral replication and progression of the disease. Sirt2 diversely affects HBV replication such that its isoform 1 intensely augments HBV replication and isoform 2 (despite of the truncated N-terminal region) moderately enhances HBV replication. Isoform 5, on the other hand, tends to protect the cell (for smooth long-term continued viral replication) from HBV-induced extreme damage or death via a discrete set of regulatory mechanisms impeding viral mRNAs, the hepatitis B core/capsid protein (HBc), core particles, replicative intermediate (RI) DNAs, and covalently closed circular DNA (cccDNA) levels, and, consequently, limiting HBV replication. In contrast to Sirt2.1 and Sirt 2.2, the Sirt2.5-mediated HBV replication is independent of the AKT/GSK3β/β-catenin signaling cascade. Sirt2.5 is recruited more at cccDNA than the recruitment of Sirt2.1 onto the cccDNA. This recruitment causes the deposition of more histone lysine methyltransferases (HKMTs), including SETDB1, SUV39H1, EZH2, and PR-Set7, along with the respective corresponding transcriptional repressive markers such as H3K9me3, H3K27me3, and H4K20me1 onto the HBV cccDNA. In HBV-replicating cells, Sirt2.5 can also make complexes with PR-Set7 and SETDB1. In addition, Sirt2.5 has the ability to turn off transcription from cccDNA through epigenetic modification via either direct or indirect interaction with HKMTs.
Keywords: HBV replication; Sirt2.1; Sirt2.2; Sirt2.5; sirtuin 2; sirtuins.
Copyright © 2024 Piracha, Saeed, Piracha, Noor and Noor.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
An Alternatively Spliced Sirtuin 2 Isoform 5 Inhibits Hepatitis B Virus Replication from cccDNA by Repressing Epigenetic Modifications Made by Histone Lysine Methyltransferases.J Virol. 2020 Jul 30;94(16):e00926-20. doi: 10.1128/JVI.00926-20. Print 2020 Jul 30. J Virol. 2020. PMID: 32493816 Free PMC article.
-
Sirtuin 2 Isoform 1 Enhances Hepatitis B Virus RNA Transcription and DNA Synthesis through the AKT/GSK-3β/β-Catenin Signaling Pathway.J Virol. 2018 Oct 12;92(21):e00955-18. doi: 10.1128/JVI.00955-18. Print 2018 Nov 1. J Virol. 2018. PMID: 30111572 Free PMC article.
-
SIRT2 Promotes HBV Transcription and Replication by Targeting Transcription Factor p53 to Increase the Activities of HBV Enhancers and Promoters.Front Microbiol. 2022 May 19;13:836446. doi: 10.3389/fmicb.2022.836446. eCollection 2022. Front Microbiol. 2022. PMID: 35663860 Free PMC article.
-
Early Steps of Hepatitis B Life Cycle: From Capsid Nuclear Import to cccDNA Formation.Viruses. 2021 Apr 26;13(5):757. doi: 10.3390/v13050757. Viruses. 2021. PMID: 33925977 Free PMC article. Review.
-
Hepatitis B Virus Capsid: The Core in Productive Entry and Covalently Closed Circular DNA Formation.Viruses. 2023 Feb 28;15(3):642. doi: 10.3390/v15030642. Viruses. 2023. PMID: 36992351 Free PMC article. Review.
References
-
- Belloni L., Allweiss L., Guerrieri F., Pediconi N., Volz T., Pollicino T., et al. . (2012). IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J. Clin. Investig. 122, 529–5537. doi: 10.1172/JCI58847 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

