Ginsenoside 20(S)-Rg3 reduces KIF20A expression and promotes CDC25A proteasomal degradation in epithelial ovarian cancer
- PMID: 38223825
- PMCID: PMC10785255
- DOI: 10.1016/j.jgr.2023.06.008
Ginsenoside 20(S)-Rg3 reduces KIF20A expression and promotes CDC25A proteasomal degradation in epithelial ovarian cancer
Abstract
Background: Ginsenoside 20(S)-Rg3 shows promising tumor-suppressive effects in ovarian cancer via inhibiting NF-κB signaling. This study aimed to explore the downstream tumor suppressive mechanisms of ginsenoside Rg3 via this signaling pathway.
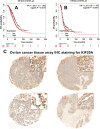
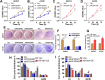
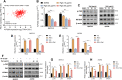
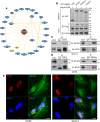
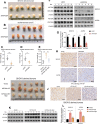
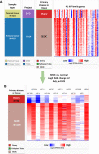
Materials and methods: A systematical screening was applied to examine the expression profile of 41 kinesin family member genes in ovarian cancer. The regulatory effect of ginsenoside Rg3 on KIF20A expression was studied. In addition, we explored interacting proteins of KIF20A and their molecular regulations in ovarian cancer. RNA-seq data from The Cancer Genome Atlas (TCGA) was used for bioinformatic analysis. Epithelial ovarian cancer cell lines SKOV3 and A2780 were used as in vitro and in vivo cell models. Commercial human ovarian cancer tissue arrays were used for immunohistochemistry staining.
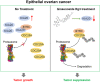
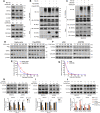
Results: KIF20A is a biomarker of poor prognosis among the kinesin genes. It promotes ovarian cancer cell growth in vitro and in vivo. Ginsenoside Rg3 can suppress the transcription of KIF20A. GST pull-down and co-immunoprecipitation (IP) assays confirmed that KIF20A physically interacts with BTRC (β-TrCP1), a substrate recognition subunit for SCFβ-TrCP E3 ubiquitin ligase. In vitro ubiquitination and cycloheximide (CHX) chase assays showed that via interacting with BTRC, KIF20A reduces BTRC-mediated CDC25A poly-ubiquitination and enhances its stability. Ginsenoside Rg3 treatment partly abrogates KIF20A overexpression-induced CDC25A upregulation.
Conclusion: This study revealed a novel anti-tumor mechanism of ginsenoside Rg3. It can inhibit KIF20A transcription and promote CDC25A proteasomal degradation in epithelial ovarian cancer.
Keywords: CDC25A; KIF20A; ginsenoside Rg3; ovarian cancer; ubiquitination.
© 2023 The Korean Society of Ginseng. Publishing services by Elsevier B.V.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Ginsenoside Rg3 increases gemcitabine sensitivity of pancreatic adenocarcinoma via reducing ZFP91 mediated TSPYL2 destabilization.J Ginseng Res. 2022 Sep;46(5):636-645. doi: 10.1016/j.jgr.2021.08.004. Epub 2021 Aug 30. J Ginseng Res. 2022. PMID: 36090681 Free PMC article.
-
Ginsenoside 20(S)-Rg3 Inhibits the Warburg Effect Via Modulating DNMT3A/ MiR-532-3p/HK2 Pathway in Ovarian Cancer Cells.Cell Physiol Biochem. 2018;45(6):2548-2559. doi: 10.1159/000488273. Epub 2018 Mar 16. Cell Physiol Biochem. 2018. PMID: 29558748
-
Ginsenoside 20(S)-Rg3 Prevents PKM2-Targeting miR-324-5p from H19 Sponging to Antagonize the Warburg Effect in Ovarian Cancer Cells.Cell Physiol Biochem. 2018;51(3):1340-1353. doi: 10.1159/000495552. Epub 2018 Nov 27. Cell Physiol Biochem. 2018. PMID: 30481782
-
Anti-Angiogenic Properties of Ginsenoside Rg3.Molecules. 2020 Oct 23;25(21):4905. doi: 10.3390/molecules25214905. Molecules. 2020. PMID: 33113992 Free PMC article. Review.
-
Advancing Cancer Therapy: The Role of KIF20A as a Target for Inhibitor Development and Immunotherapy.Cancers (Basel). 2024 Aug 24;16(17):2958. doi: 10.3390/cancers16172958. Cancers (Basel). 2024. PMID: 39272816 Free PMC article. Review.
Cited by
-
A nanoscale natural drug delivery system for targeted drug delivery against ovarian cancer: action mechanism, application enlightenment and future potential.Front Immunol. 2024 Oct 11;15:1427573. doi: 10.3389/fimmu.2024.1427573. eCollection 2024. Front Immunol. 2024. PMID: 39464892 Free PMC article. Review.
References
-
- Crean-Tate K.K., Braley C., Dey G., Esakov E., Saygin C., Trestan A., et al. Pretreatment with LCK inhibitors chemosensitizes cisplatin-resistant endometrioid ovarian tumors. J Ovarian Res. 2021;14(1):55. doi: 10.1186/s13048-021-00797-x. Epub 2021/04/24. PubMed PMID: 33888137; PubMed Central PMCID: PMCPMC8063392. - DOI - PMC - PubMed
-
- Franzese E., Diana A., Centonze S., Pignata S., De Vita F., Ciardiello F., et al. PARP Inhibitors in First-Line Therapy of Ovarian Cancer: Are There Any Doubts? Front Oncol. 2020;10:782. doi: 10.3389/fonc.2020.00782. Epub 2020/07/01. PubMed PMID: 32596142; PubMed Central PMCID: PMCPMC7303974. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

