Transcription factor FoxO1 regulates myoepithelial cell diversity and growth
- PMID: 38212454
- PMCID: PMC10784559
- DOI: 10.1038/s41598-024-51619-1
Transcription factor FoxO1 regulates myoepithelial cell diversity and growth
Abstract
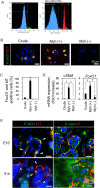
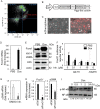
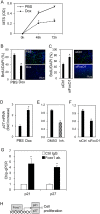
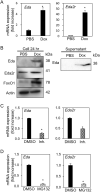
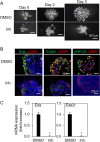
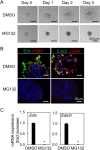
Salivary gland myoepithelial cells regulate saliva secretion and have been implicated in the histological diversity of salivary gland tumors. However, detailed functional analysis of myoepithelial cells has not been determined owing to the few of the specific marker to isolate them. We isolated myoepithelial cells from the submandibular glands of adult mice using the epithelial marker EpCAM and the cell adhesion molecule CD49f as indicators and found predominant expression of the transcription factor FoxO1 in these cells. RNA-sequence analysis revealed that the expression of cell cycle regulators was negatively regulated in FoxO1-overexpressing cells. Chromatin immunoprecipitation analysis showed that FoxO1 bound to the p21/p27 promoter DNA, indicating that FoxO1 suppresses cell proliferation through these factors. In addition, FoxO1 induced the expression of ectodysplasin A (Eda) and its receptor Eda2r, which are known to be associated with X-linked hypohidrotic ectodermal dysplasia and are involved in salivary gland development in myoepithelial cells. FoxO1 inhibitors suppressed Eda/Eda2r expression and salivary gland development in primordial organ cultures after mesenchymal removal. Although mesenchymal cells are considered a source of Eda, myoepithelial cells might be one of the resources of Eda. These results suggest that FoxO1 regulates myoepithelial cell proliferation and Eda secretion during salivary gland development in myoepithelial cells.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Defective NaCl Reabsorption in Salivary Glands of Eda-Null X-LHED Mice.J Dent Res. 2018 Oct;97(11):1244-1251. doi: 10.1177/0022034518782461. Epub 2018 Jun 18. J Dent Res. 2018. PMID: 29913094 Free PMC article.
-
Isolation and Functional Analysis of Myoepithelial Cells from Adult Mouse Submandibular Glands.Methods Mol Biol. 2024;2736:53-64. doi: 10.1007/7651_2022_472. Methods Mol Biol. 2024. PMID: 36749482
-
Ectodysplasin-A2 induces apoptosis in cultured human hair follicle cells and promotes regression of hair follicles in mice.Biochem Biophys Res Commun. 2019 Dec 3;520(2):428-433. doi: 10.1016/j.bbrc.2019.10.031. Epub 2019 Oct 11. Biochem Biophys Res Commun. 2019. PMID: 31607478
-
Ectodysplasin signaling in development.Cytokine Growth Factor Rev. 2003 Jun-Aug;14(3-4):211-24. doi: 10.1016/s1359-6101(03)00020-0. Cytokine Growth Factor Rev. 2003. PMID: 12787560 Review.
-
Ectodysplasin A (EDA) - EDA receptor signalling and its pharmacological modulation.Cytokine Growth Factor Rev. 2014 Apr;25(2):195-203. doi: 10.1016/j.cytogfr.2014.01.004. Epub 2014 Jan 23. Cytokine Growth Factor Rev. 2014. PMID: 24508088 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

