Genomic analysis of diverse environmental Acinetobacter isolates identifies plasmids, antibiotic resistance genes, and capsular polysaccharides shared with clinical strains
- PMID: 38206028
- PMCID: PMC10885009
- DOI: 10.1128/aem.01654-23
Genomic analysis of diverse environmental Acinetobacter isolates identifies plasmids, antibiotic resistance genes, and capsular polysaccharides shared with clinical strains
Abstract
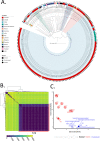
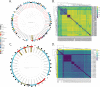
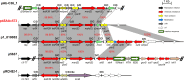
Acinetobacter baumannii, an important pathogen known for its widespread antibiotic resistance, has been the focus of extensive research within its genus, primarily involving clinical isolates. Consequently, data on environmental A. baumannii and other Acinetobacter species remain limited. Here, we utilized Illumina and Nanopore sequencing to analyze the genomes of 10 Acinetobacter isolates representing 6 different species sourced from aquatic environments in South Australia. All 10 isolates were phylogenetically distinct compared to clinical and other non-clinical Acinetobacter strains, often tens of thousands of single-nucleotide polymorphisms from their nearest neighbors. Despite the genetic divergence, we identified pdif modules (sections of mobilized DNA) carrying clinically important antimicrobial resistance genes in species other than A. baumannii, including carbapenemase oxa58, tetracycline resistance gene tet(39), and macrolide resistance genes msr(E)-mph(E). These pdif modules were located on plasmids with high sequence identity to those circulating in globally distributed A. baumannii ST1 and ST2 clones. The environmental A. baumannii isolate characterized here (SAAb472; ST350) did not possess any native plasmids; however, it could capture two clinically important plasmids (pRAY and pACICU2) with high transfer frequencies. Furthermore, A. baumannii SAAb472 possessed virulence genes and a capsular polysaccharide type analogous to clinical strains. Our findings highlight the potential for environmental Acinetobacter species to acquire and disseminate clinically important antimicrobial resistance genes, underscoring the need for further research into the ecology and evolution of this important genus.IMPORTANCEAntimicrobial resistance (AMR) is a global threat to human, animal, and environmental health. Studying AMR in environmental bacteria is crucial to understand the emergence and dissemination of resistance genes and pathogens, and to identify potential reservoirs and transmission routes. This study provides novel insights into the genomic diversity and AMR potential of environmental Acinetobacter species. By comparing the genomes of aquatic Acinetobacter isolates with clinical and non-clinical strains, we revealed that they are highly divergent yet carry pdif modules that encode resistance to antibiotics commonly used in clinical settings. We also demonstrated that an environmental A. baumannii isolate can acquire clinically relevant plasmids and carries virulence factors similar to those of hospital-associated strains. These findings suggest that environmental Acinetobacter species may serve as reservoirs and vectors of clinically important genes. Consequently, further research is warranted to comprehensively understand the ecology and evolution of this genus.
Keywords: Acinetobacter baumannii; Acinetobacter chinensis; Acinetobacter gerneri; Acinetobacter johnsonii; Acinetobacter towneri; antibiotic resistance; environmental; mobile genetic elements; plasmid; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Genomic Analysis of Carbapenem-Resistant Acinetobacter baumannii Strains Recovered from Chilean Hospitals Reveals Lineages Specific to South America and Multiple Routes for Acquisition of Antibiotic Resistance Genes.Microbiol Spectr. 2022 Oct 26;10(5):e0246322. doi: 10.1128/spectrum.02463-22. Epub 2022 Sep 26. Microbiol Spectr. 2022. PMID: 36154439 Free PMC article.
-
Accumulation of Antibiotic Resistance Genes in Carbapenem-Resistant Acinetobacter baumannii Isolates Belonging to Lineage 2, Global Clone 1, from Outbreaks in 2012-2013 at a Tehran Burns Hospital.mSphere. 2020 Apr 8;5(2):e00164-20. doi: 10.1128/mSphere.00164-20. mSphere. 2020. PMID: 32269158 Free PMC article.
-
Complete Genome Sequencing of Acinetobacter baumannii AC1633 and Acinetobacter nosocomialis AC1530 Unveils a Large Multidrug-Resistant Plasmid Encoding the NDM-1 and OXA-58 Carbapenemases.mSphere. 2021 Jan 27;6(1):e01076-20. doi: 10.1128/mSphere.01076-20. mSphere. 2021. PMID: 33504662 Free PMC article.
-
What do we know about plasmids carried by members of the Acinetobacter genus?World J Microbiol Biotechnol. 2020 Jul 13;36(8):109. doi: 10.1007/s11274-020-02890-7. World J Microbiol Biotechnol. 2020. PMID: 32656745 Review.
-
Mobilization of pdif modules in Acinetobacter: A novel mechanism for antibiotic resistance gene shuffling?Mol Microbiol. 2020 Nov;114(5):699-709. doi: 10.1111/mmi.14563. Epub 2020 Jul 21. Mol Microbiol. 2020. PMID: 32594594 Review.
Cited by
-
Decoding the resistome, virulome and mobilome of clinical versus aquatic Acinetobacter baumannii in southern Romania.Heliyon. 2024 Jun 21;10(13):e33372. doi: 10.1016/j.heliyon.2024.e33372. eCollection 2024 Jul 15. Heliyon. 2024. PMID: 39035534 Free PMC article.
-
A Review on Colistin Resistance: An Antibiotic of Last Resort.Microorganisms. 2024 Apr 11;12(4):772. doi: 10.3390/microorganisms12040772. Microorganisms. 2024. PMID: 38674716 Free PMC article. Review.
-
Carbapenemase genes in clinical and environmental isolates of Acinetobacter spp. from Quito, Ecuador.PeerJ. 2024 Apr 25;12:e17199. doi: 10.7717/peerj.17199. eCollection 2024. PeerJ. 2024. PMID: 38680892 Free PMC article.
-
Genomic Analysis of Antimicrobial Resistance in Pseudomonas aeruginosa from a "One Health" Perspective.Microorganisms. 2024 Aug 27;12(9):1770. doi: 10.3390/microorganisms12091770. Microorganisms. 2024. PMID: 39338445 Free PMC article.
References
-
- Koong J, Johnson C, Rafei R, Hamze M, Myers GSA, Kenyon JJ, Lopatkin AJ, Hamidian M. 2021. Phylogenomics of two ST1 antibiotic-susceptible non-clinical Acinetobacter baumannii strains reveals multiple lineages and complex evolutionary history in global clone 1. Microb Genom 7:000705. doi:10.1099/mgen.0.000705 - DOI - PMC - PubMed
-
- Furlan JPR, de Almeida OGG, De Martinis ECP, Stehling EG. 2019. Characterization of an environmental multidrug-resistant Acinetobacter seifertii and comparative genomic analysis reveals co-occurrence of antimicrobial resistance and metal tolerance determinants. Front Microbiol 10:2151. doi:10.3389/fmicb.2019.02151 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

