Characterization of Tau95 led to the identification of a four-subunit TFIIIC complex in trypanosomatid parasites
- PMID: 38204130
- PMCID: PMC10781861
- DOI: 10.1007/s00253-023-12903-8
Characterization of Tau95 led to the identification of a four-subunit TFIIIC complex in trypanosomatid parasites
Abstract
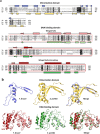
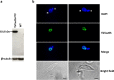
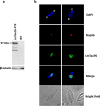
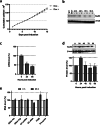
RNA polymerase III (RNAP III) synthetizes small essential non-coding RNA molecules such as tRNAs and 5S rRNA. In yeast and vertebrates, RNAP III needs general transcription factors TFIIIA, TFIIIB, and TFIIIC to initiate transcription. TFIIIC, composed of six subunits, binds to internal promoter elements in RNAP III-dependent genes. Limited information is available about RNAP III transcription in the trypanosomatid protozoa Trypanosoma brucei and Leishmania major, which diverged early from the eukaryotic lineage. Analyses of the first published draft of the trypanosomatid genome sequences failed to recognize orthologs of any of the TFIIIC subunits, suggesting that this transcription factor is absent in these parasites. However, a putative TFIIIC subunit was recently annotated in the databases. Here we characterize this subunit in T. brucei and L. major and demonstrate that it corresponds to Tau95. In silico analyses showed that both proteins possess the typical Tau95 sequences: the DNA binding region and the dimerization domain. As anticipated for a transcription factor, Tau95 localized to the nucleus in insect forms of both parasites. Chromatin immunoprecipitation (ChIP) assays demonstrated that Tau95 binds to tRNA and U2 snRNA genes in T. brucei. Remarkably, by performing tandem affinity purifications we identified orthologs of TFIIIC subunits Tau55, Tau131, and Tau138 in T. brucei and L. major. Thus, contrary to what was assumed, trypanosomatid parasites do possess a TFIIIC complex. Other putative interacting partners of Tau95 were identified in T. brucei and L. major. KEY POINTS: • A four-subunit TFIIIC complex is present in T. brucei and L. major • TbTau95 associates with tRNA and U2 snRNA genes • Putative interacting partners of Tau95 might include some RNAP II regulators.
Keywords: 5S rRNA; Leishmania major; RNAP III transcription; TFIIIC; Tau95; Trypanosoma brucei; tRNA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Analyses of the essential C82 subunit uncovered some differences in RNA polymerase III transcription between Trypanosoma brucei and Leishmania major.Parasitology. 2024 Nov 11:1-16. doi: 10.1017/S0031182024000921. Online ahead of print. Parasitology. 2024. PMID: 39523652
-
The tau95 subunit of yeast TFIIIC influences upstream and downstream functions of TFIIIC.DNA complexes.J Biol Chem. 2003 Mar 21;278(12):10450-7. doi: 10.1074/jbc.M213310200. Epub 2003 Jan 16. J Biol Chem. 2003. PMID: 12533520
-
Participation of TFIIIB Subunit Brf1 in Transcription Regulation in the Human Pathogen Leishmania major.Genes (Basel). 2021 Feb 16;12(2):280. doi: 10.3390/genes12020280. Genes (Basel). 2021. PMID: 33669344 Free PMC article.
-
Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human.Nucleic Acids Res. 2001 Jul 1;29(13):2675-90. doi: 10.1093/nar/29.13.2675. Nucleic Acids Res. 2001. PMID: 11433012 Free PMC article. Review.
-
Regulation of tRNA synthesis by the general transcription factors of RNA polymerase III - TFIIIB and TFIIIC, and by the MAF1 protein.Biochim Biophys Acta Gene Regul Mech. 2018 Apr;1861(4):320-329. doi: 10.1016/j.bbagrm.2018.01.011. Epub 2018 Feb 6. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 29378333 Review.
References
-
- Billington K, Halliday C, Madden R, Dyer P, Barker AR, Moreira-Leite FF, Carrington M, Vaughan S, Hertz-Fowler C, Dean S, Sunter JD, Wheeler RJ, Gull K. Genome-wide subcellular protein map for the flagellate parasite Trypanosoma brucei. Nat Microbiol. 2023;8(3):533–547. doi: 10.1038/s41564-022-01295-6. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

