OsNAC103, an NAC transcription factor negatively regulates plant height in rice
- PMID: 38193994
- PMCID: PMC10776745
- DOI: 10.1007/s00425-023-04309-7
OsNAC103, an NAC transcription factor negatively regulates plant height in rice
Abstract
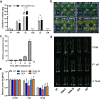
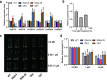
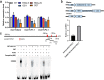
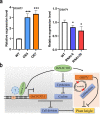
OsNAC103 negatively regulates rice plant height by influencing the cell cycle and crosstalk of phytohormones. Plant height is an important characteristic of rice farming and is directly related to agricultural yield. Although there has been great progress in research on plant growth regulation, numerous genes remain to be elucidated. NAC transcription factors are widespread in plants and have a vital function in plant growth. Here, we observed that the overexpression of OsNAC103 resulted in a dwarf phenotype, whereas RNA interference (RNAi) plants and osnac103 mutants showed no significant difference. Further investigation revealed that the cell length did not change, indicating that the dwarfing of plants was caused by a decrease in cell number due to cell cycle arrest. The content of the bioactive cytokinin N6-Δ2-isopentenyladenine (iP) decreased as a result of the cytokinin synthesis gene being downregulated and the enhanced degradation of cytokinin oxidase. OsNAC103 overexpression also inhibited cell cycle progression and regulated the activity of the cell cyclin OsCYCP2;1 to arrest the cell cycle. We propose that OsNAC103 may further influence rice development and gibberellin-cytokinin crosstalk by regulating the Oryza sativa homeobox 71 (OSH71). Collectively, these results offer novel perspectives on the role of OsNAC103 in controlling plant architecture.
Keywords: Cell cycle; Cytokinins; Gibberellins; Phytohormones crosstalk; Plant development.
© 2024. The Author(s).
Conflict of interest statement
No competing or financial interests were declared.
Figures


Similar articles
-
OsNAC103, a NAC Transcription Factor, Positively Regulates Leaf Senescence and Plant Architecture in Rice.Rice (N Y). 2024 Feb 15;17(1):15. doi: 10.1186/s12284-024-00690-3. Rice (N Y). 2024. PMID: 38358523 Free PMC article.
-
Strigolactone promotes cytokinin degradation through transcriptional activation of CYTOKININ OXIDASE/DEHYDROGENASE 9 in rice.Proc Natl Acad Sci U S A. 2019 Jul 9;116(28):14319-14324. doi: 10.1073/pnas.1810980116. Epub 2019 Jun 24. Proc Natl Acad Sci U S A. 2019. PMID: 31235564 Free PMC article.
-
Dwarf Tiller1, a Wuschel-related homeobox transcription factor, is required for tiller growth in rice.PLoS Genet. 2014 Mar 13;10(3):e1004154. doi: 10.1371/journal.pgen.1004154. eCollection 2014 Mar. PLoS Genet. 2014. PMID: 24625559 Free PMC article.
-
Phytohormones and rice crop yield: strategies and opportunities for genetic improvement.Transgenic Res. 2006 Aug;15(4):399-404. doi: 10.1007/s11248-006-0024-1. Transgenic Res. 2006. PMID: 16906440 Review.
-
The genetic and molecular basis of crop height based on a rice model.Planta. 2018 Jan;247(1):1-26. doi: 10.1007/s00425-017-2798-1. Epub 2017 Nov 6. Planta. 2018. PMID: 29110072 Review.
Cited by
-
Transcriptional Control of Seed Life: New Insights into the Role of the NAC Family.Int J Mol Sci. 2024 May 14;25(10):5369. doi: 10.3390/ijms25105369. Int J Mol Sci. 2024. PMID: 38791407 Free PMC article. Review.
References
-
- Camut L, Gallova B, Jilli L, Sirlin-Josserand M, Carrera E, Sakvarelidze-Achard L, Ruffel S, Krouk G, Thomas SG, Hedden P, Phillips AL, Daviere JM, Achard P. Nitrate signaling promotes plant growth by upregulating gibberellin biosynthesis and destabilization of DELLA proteins. Curr Biol. 2021;31(22):4971–4982, e4974. doi: 10.1016/j.cub.2021.09.024. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

