Identifying patterns to uncover the importance of biological pathways on known drug repurposing scenarios
- PMID: 38191292
- PMCID: PMC10775474
- DOI: 10.1186/s12864-023-09913-1
Identifying patterns to uncover the importance of biological pathways on known drug repurposing scenarios
Abstract
Background: Drug repurposing plays a significant role in providing effective treatments for certain diseases faster and more cost-effectively. Successful repurposing cases are mostly supported by a classical paradigm that stems from de novo drug development. This paradigm is based on the "one-drug-one-target-one-disease" idea. It consists of designing drugs specifically for a single disease and its drug's gene target. In this article, we investigated the use of biological pathways as potential elements to achieve effective drug repurposing.
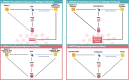
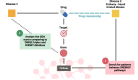
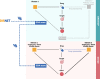
Methods: Considering a total of 4214 successful cases of drug repurposing, we identified cases in which biological pathways serve as the underlying basis for successful repurposing, referred to as DREBIOP. Once the repurposing cases based on pathways were identified, we studied their inherent patterns by considering the different biological elements associated with this dataset, as well as the pathways involved in these cases. Furthermore, we obtained gene-disease association values to demonstrate the diminished significance of the drug's gene target in these repurposing cases. To achieve this, we compared the values obtained for the DREBIOP set with the overall association values found in DISNET, as well as with the drug's target gene (DREGE) based repurposing cases using the Mann-Whitney U Test.
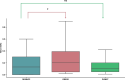
Results: A collection of drug repurposing cases, known as DREBIOP, was identified as a result. DREBIOP cases exhibit distinct characteristics compared with DREGE cases. Notably, DREBIOP cases are associated with a higher number of biological pathways, with Vitamin D Metabolism and ACE inhibitors being the most prominent pathways. Additionally, it was observed that the association values of GDAs in DREBIOP cases were significantly lower than those in DREGE cases (p-value < 0.05).
Conclusions: Biological pathways assume a pivotal role in drug repurposing cases. This investigation successfully revealed patterns that distinguish drug repurposing instances associated with biological pathways. These identified patterns can be applied to any known repurposing case, enabling the detection of pathway-based repurposing scenarios or the classical paradigm.
Keywords: Computational biology; DISNET knowledge; Data-driven methodology; Drug repurposing; Pathway-based.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
A data-driven methodology towards evaluating the potential of drug repurposing hypotheses.Comput Struct Biotechnol J. 2021 Aug 9;19:4559-4573. doi: 10.1016/j.csbj.2021.08.003. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34471499 Free PMC article.
-
Repurposing old molecules for new indications: Defining pillars of success from lessons in the past.Eur J Pharmacol. 2021 Dec 5;912:174569. doi: 10.1016/j.ejphar.2021.174569. Epub 2021 Oct 13. Eur J Pharmacol. 2021. PMID: 34653378 Review.
-
Protein sequence analysis in the context of drug repurposing.BMC Med Inform Decis Mak. 2024 May 13;24(1):122. doi: 10.1186/s12911-024-02531-1. BMC Med Inform Decis Mak. 2024. PMID: 38741115 Free PMC article.
-
Drug repurposing for Parkinson's disease by biological pathway based edge-weighted network proximity analysis.Sci Rep. 2024 Sep 11;14(1):21258. doi: 10.1038/s41598-024-71922-1. Sci Rep. 2024. PMID: 39261542 Free PMC article.
-
Computer-aided drug repurposing for cancer therapy: Approaches and opportunities to challenge anticancer targets.Semin Cancer Biol. 2021 Jan;68:59-74. doi: 10.1016/j.semcancer.2019.09.023. Epub 2019 Sep 25. Semin Cancer Biol. 2021. PMID: 31562957 Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

