N-terminal acetyltransferase 6 facilitates enterovirus 71 replication by regulating PI4KB expression and replication organelle biogenesis
- PMID: 38189249
- PMCID: PMC10878262
- DOI: 10.1128/jvi.01749-23
N-terminal acetyltransferase 6 facilitates enterovirus 71 replication by regulating PI4KB expression and replication organelle biogenesis
Abstract
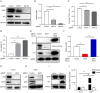
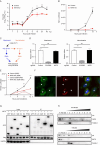
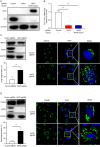
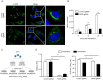
Enterovirus 71 (EV71) is one of the major pathogens causing hand, foot, and mouth disease in children under 5 years old, which can result in severe neurological complications and even death. Due to limited treatments for EV71 infection, the identification of novel host factors and elucidation of mechanisms involved will help to counter this viral infection. N-terminal acetyltransferase 6 (NAT6) was identified as an essential host factor for EV71 infection with genome-wide CRISPR/Cas9 screening. NAT6 facilitates EV71 viral replication depending on its acetyltransferase activity but has little effect on viral release. In addition, NAT6 is also required for Echovirus 7 and coxsackievirus B5 infection, suggesting it might be a pan-enterovirus host factor. We further demonstrated that NAT6 is required for Golgi integrity and viral replication organelle (RO) biogenesis. NAT6 knockout significantly inhibited phosphatidylinositol 4-kinase IIIβ (PI4KB) expression and PI4P production, both of which are key host factors for enterovirus infection and RO biogenesis. Further mechanism studies confirmed that NAT6 formed a complex with its substrate actin and one of the PI4KB recruiters-acyl-coenzyme A binding domain containing 3 (ACBD3). Through modulating actin dynamics, NAT6 maintained the integrity of the Golgi and the stability of ACBD3, thereby enhancing EV71 infection. Collectively, these results uncovered a novel mechanism of N-acetyltransferase supporting EV71 infection.IMPORTANCEEnterovirus 71 (EV71) is an important pathogen for children under the age of five, and currently, no effective treatment is available. Elucidating the mechanism of novel host factors supporting viral infection will reveal potential antiviral targets and aid antiviral development. Here, we demonstrated that a novel N-acetyltransferase, NAT6, is an essential host factor for EV71 replication. NAT6 could promote viral replication organelle (RO) formation to enhance viral replication. The formation of enterovirus ROs requires numerous host factors, including acyl-coenzyme A binding domain containing 3 (ACBD3) and phosphatidylinositol 4-kinase IIIβ (PI4KB). NAT6 could stabilize the PI4KB recruiter, ACBD3, by inhibiting the autophagy degradation pathway. This study provides a fresh insight into the relationship between N-acetyltransferase and viral infection.
Keywords: ACBD3; Golgi morphology; acetyltransferase; enteroviruses; replication organelle.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
ACBD3 Is an Essential Pan-enterovirus Host Factor That Mediates the Interaction between Viral 3A Protein and Cellular Protein PI4KB.mBio. 2019 Feb 12;10(1):e02742-18. doi: 10.1128/mBio.02742-18. mBio. 2019. PMID: 30755512 Free PMC article.
-
A complex comprising phosphatidylinositol 4-kinase IIIβ, ACBD3, and Aichi virus proteins enhances phosphatidylinositol 4-phosphate synthesis and is critical for formation of the viral replication complex.J Virol. 2014 Jun;88(12):6586-98. doi: 10.1128/JVI.00208-14. Epub 2014 Mar 26. J Virol. 2014. PMID: 24672044 Free PMC article.
-
Enterovirus 3A Facilitates Viral Replication by Promoting Phosphatidylinositol 4-Kinase IIIβ-ACBD3 Interaction.J Virol. 2017 Sep 12;91(19):e00791-17. doi: 10.1128/JVI.00791-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28701404 Free PMC article.
-
[Molecular Mechanism of Action of hnRNP K and RTN3 in the Replication of Enterovirus 71].Bing Du Xue Bao. 2015 Mar;31(2):197-200. Bing Du Xue Bao. 2015. PMID: 26164948 Review. Chinese.
-
Emerging Role for Acyl-CoA Binding Domain Containing 3 at Membrane Contact Sites During Viral Infection.Front Microbiol. 2020 Apr 8;11:608. doi: 10.3389/fmicb.2020.00608. eCollection 2020. Front Microbiol. 2020. PMID: 32322249 Free PMC article. Review.
Cited by
-
The class III phosphatidylinositol 3-kinase VPS34 supports EV71 replication by promoting viral replication organelle formation.J Virol. 2024 Oct 22;98(10):e0069524. doi: 10.1128/jvi.00695-24. Epub 2024 Sep 10. J Virol. 2024. PMID: 39254312
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

